| (1) HEART | (2) KIDNEY | (1) OTHER | (2) TESTES |
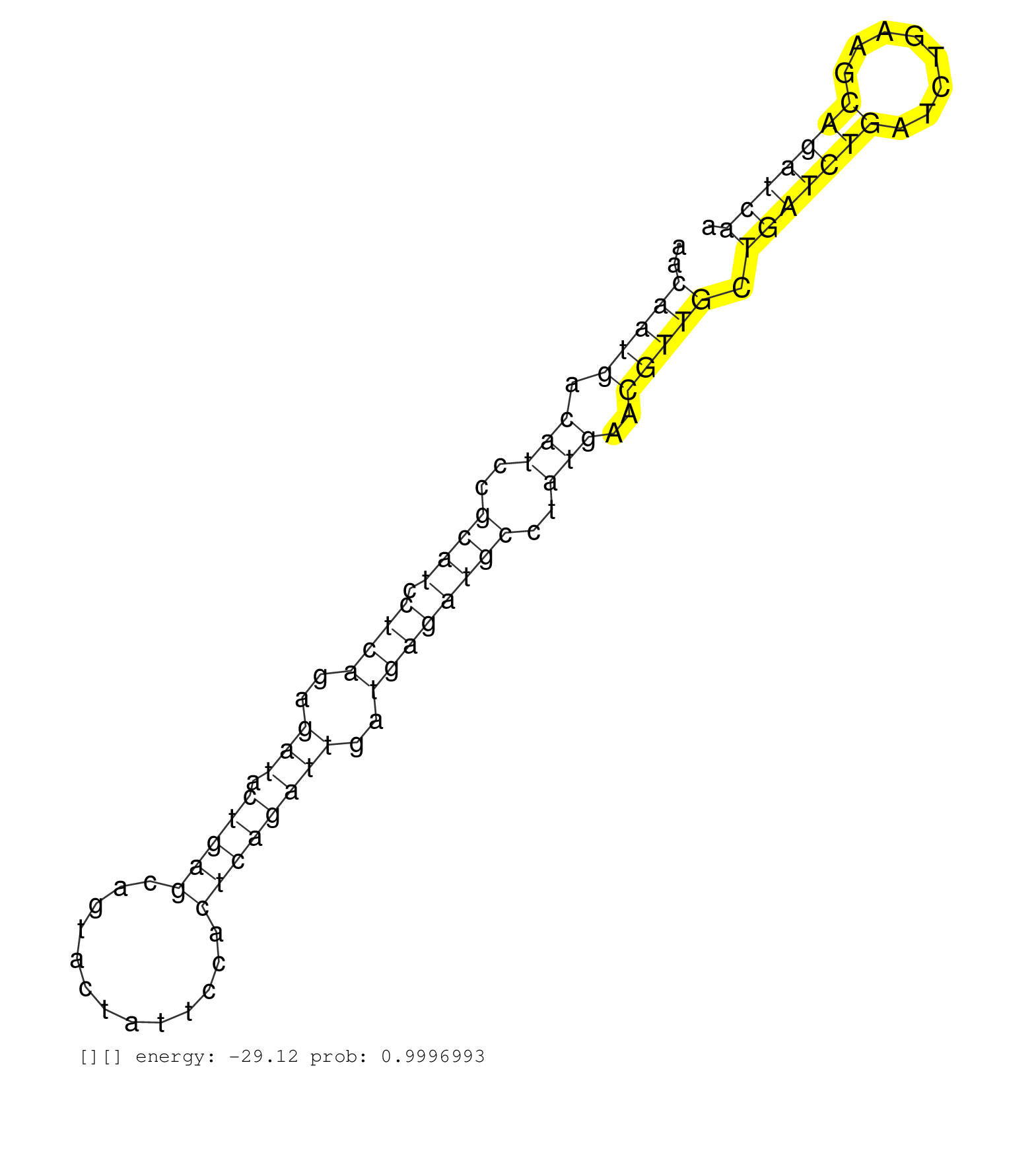
| CCCCAGAAAGAGCGGGAGTCCATCATGAAGGAGTTCCGGTCGGGCGCCAGGTGGGTCCGCTGTGCTGTGGCTTTGCCGGTCAGGGTTTGCTAGACTGTGTGCTAAGCACTTCATATGCACTGCCTGAGGCAGGCCCCAGAGCTGGGGAGATGACGAGCTCCTGCTCTGAAAGCTGAGGTGTCGCCATTAACCTTGTAAAGAACAATGACATCCGCATCCTCAGAGATACTGAGCAGTACTATTCCACTCAGATTGATGAGATGCCTATGAACGTTGCTGATCTGATCTGAAGCAGATCAATGGGATGAGTGGAGACTGTTCACCTGTTCTGTACTCCTGTTTGGAAGTAT ..........................................................................................................................................................................................................(((((.(((..((((.((((..(((.(((((.............))))))))..))))))))..)))..))))).(((((((........)))))))................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553607(SRX182813) source: Testis. (testes) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|---|---|
| ......................TCATGAAGGAGTTCCGGTCGGGCGCCAGT........................................................................................................................................................................................................................................................................................................... | 29 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | |
| .........................................................................................................................................................................................................................................................................TATGAACGTTGCTGATCTGATCTGAAGCA........................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - |
| .........................................................................................................................................................................................................................................................................TATGAACGTTGCTGATCTGATCTGAAGCAG....................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - |
| ...........................................................................................................................................................................................................................................................................TGAACGTTGCTGATCTGATCTGAAGC......................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................................................................AGACTGTTCACCTGTTCTGTACT............... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - |
| .............................................................................................................................................................................................................................................................................AACGTTGCTGATCTGATCTGAAGCA........................................................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................TGAACGTTGCTGATCTGATCTGAAGCA........................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................CCTGCTCTGAAAGCT................................................................................................................................................................................ | 15 | 19 | 0.05 | 0.05 | - | - | - | - | - | 0.05 |
| CCCCAGAAAGAGCGGGAGTCCATCATGAAGGAGTTCCGGTCGGGCGCCAGGTGGGTCCGCTGTGCTGTGGCTTTGCCGGTCAGGGTTTGCTAGACTGTGTGCTAAGCACTTCATATGCACTGCCTGAGGCAGGCCCCAGAGCTGGGGAGATGACGAGCTCCTGCTCTGAAAGCTGAGGTGTCGCCATTAACCTTGTAAAGAACAATGACATCCGCATCCTCAGAGATACTGAGCAGTACTATTCCACTCAGATTGATGAGATGCCTATGAACGTTGCTGATCTGATCTGAAGCAGATCAATGGGATGAGTGGAGACTGTTCACCTGTTCTGTACTCCTGTTTGGAAGTAT ..........................................................................................................................................................................................................(((((.(((..((((.((((..(((.(((((.............))))))))..))))))))..)))..))))).(((((((........)))))))................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553607(SRX182813) source: Testis. (testes) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................................................................................................................ACAGGAGTACAGAACAGGTGAACAGTCT.......... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| .........................................................................AAACCCTGACCGGCA...................................................................................................................................................................................................................................................................... | 15 | 4 | 0.25 | 0.25 | - | - | - | - | 0.25 | - |