| (1) KIDNEY | (1) OTHER | (2) TESTES |
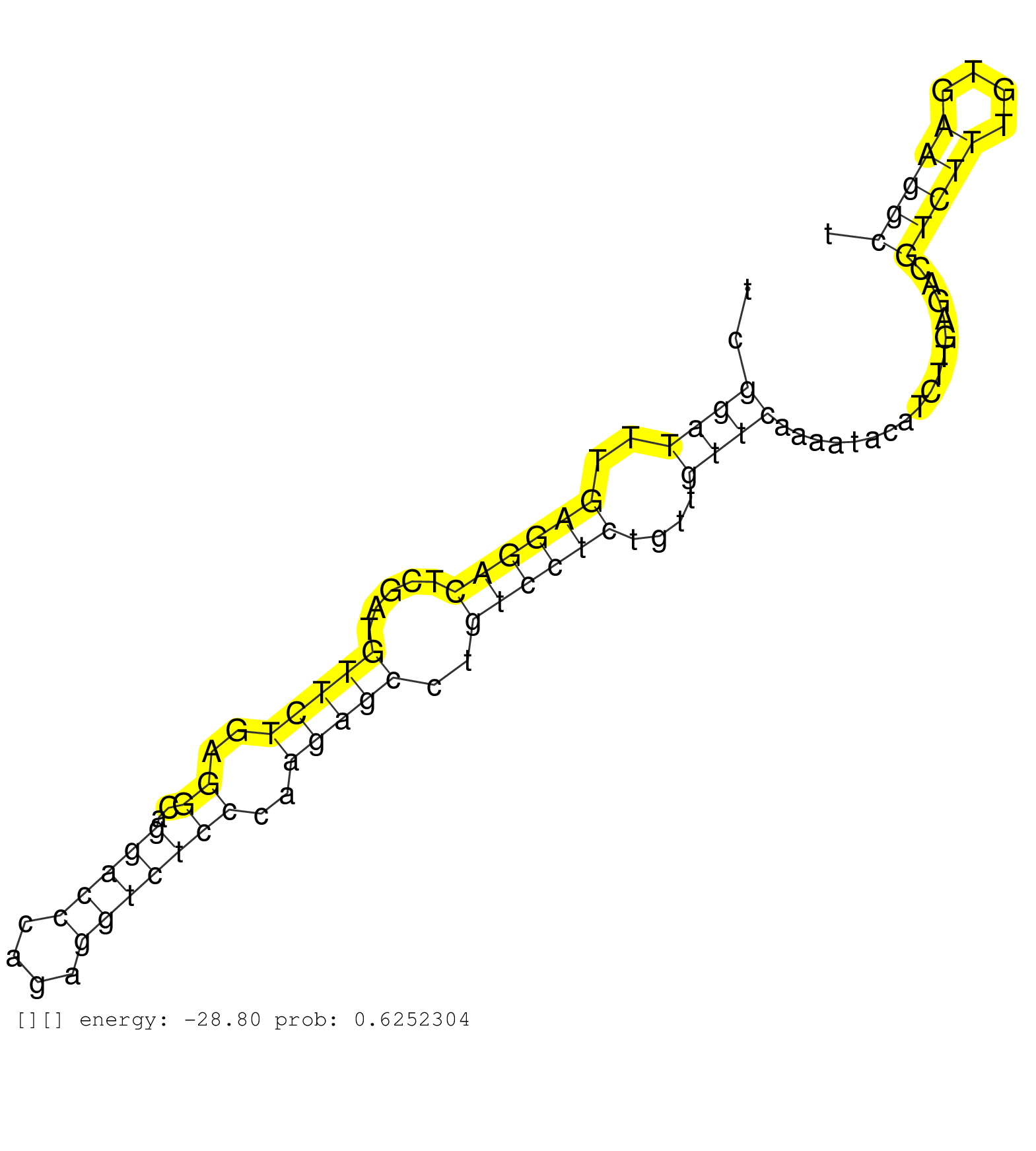
| TCCCCCCGAGGACTTCCGCGGCTTTTCCAGCCCAGCAGCAGCCGCTCCAGGTACTTTCTGATGGCTCCACAATGCAGCTCCCCAGACTTTCCTCACTCGGATTTGAGGACTCGATGTTCTGAGGCAGGACCCAGAGGTCTCCCAAGAGCCTGTCCTCTGTTGTTCAAAATACATCTTGAGACGTCTTTGTGAAGGCTCTTAGTTTTAATGCATGAATGCTGTTATTTTTCCCTACTGTTACTGAAATTAAAGTGTTTGTCTCTGATCATTAGTTCTGTTAACTTTTCTGCTGGAATGTTTTTCATTTGCATTTGCTTTTCTGATATATTTGCATTTTTCATTGCTTCTGT ..................................................................................................((((..((((((.....(((((..((..(((((....)))))))..)))))..))))))....)))).................(((((....))))).......................................................................................................................................................... ................................................................................................97..................................................................................................197....................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) | SRR553606(SRX182812) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|
| ............................................................................................TCACTCGGATTTGAGGACTCGATGTTCTT..................................................................................................................................................................................................................................... | 29 | 2.00 | 0.00 | 1.00 | 1.00 | - | - | |
| .....................................................................................................TTTGAGGACTCGATGTTCTGAGGCAGGACC........................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...................................................TACTTTCTGATGGCTCCACAATGCAGCTCC............................................................................................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .....................................................................................................TTTGAGGACTCGATGTTCTGAGGC................................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...................................................................................................................................................................TCAAAATACATCTTGAGACGTCTTTGTGAA............................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ........................................................TCTGATGGCTCCACAATGCAGCTCCCC........................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .............................................................................................................................................................................TCTTGAGACGTCTTTGTGAA............................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .............................................................................................................................................................................................TGAAGGCTCTTAGTTTTAATGCATGAATGC................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| TCCCCCCGAGGACTTCCGCGGCTTTTCCAGCCCAGCAGCAGCCGCTCCAGGTACTTTCTGATGGCTCCACAATGCAGCTCCCCAGACTTTCCTCACTCGGATTTGAGGACTCGATGTTCTGAGGCAGGACCCAGAGGTCTCCCAAGAGCCTGTCCTCTGTTGTTCAAAATACATCTTGAGACGTCTTTGTGAAGGCTCTTAGTTTTAATGCATGAATGCTGTTATTTTTCCCTACTGTTACTGAAATTAAAGTGTTTGTCTCTGATCATTAGTTCTGTTAACTTTTCTGCTGGAATGTTTTTCATTTGCATTTGCTTTTCTGATATATTTGCATTTTTCATTGCTTCTGT ..................................................................................................((((..((((((.....(((((..((..(((((....)))))))..)))))..))))))....)))).................(((((....))))).......................................................................................................................................................... ................................................................................................97..................................................................................................197....................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) | SRR553606(SRX182812) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................TGAACAACAGAGGACAG........................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ............................................................................................................................tcgaGAGACCTCTGGGTAGCT............................................................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | 1.00 | - | |
| .................................................................................................................................................AGATGTATTTTGAACAACAGAGGACAGGCTC.............................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |