| (1) KIDNEY | (2) OTHER | (2) TESTES |
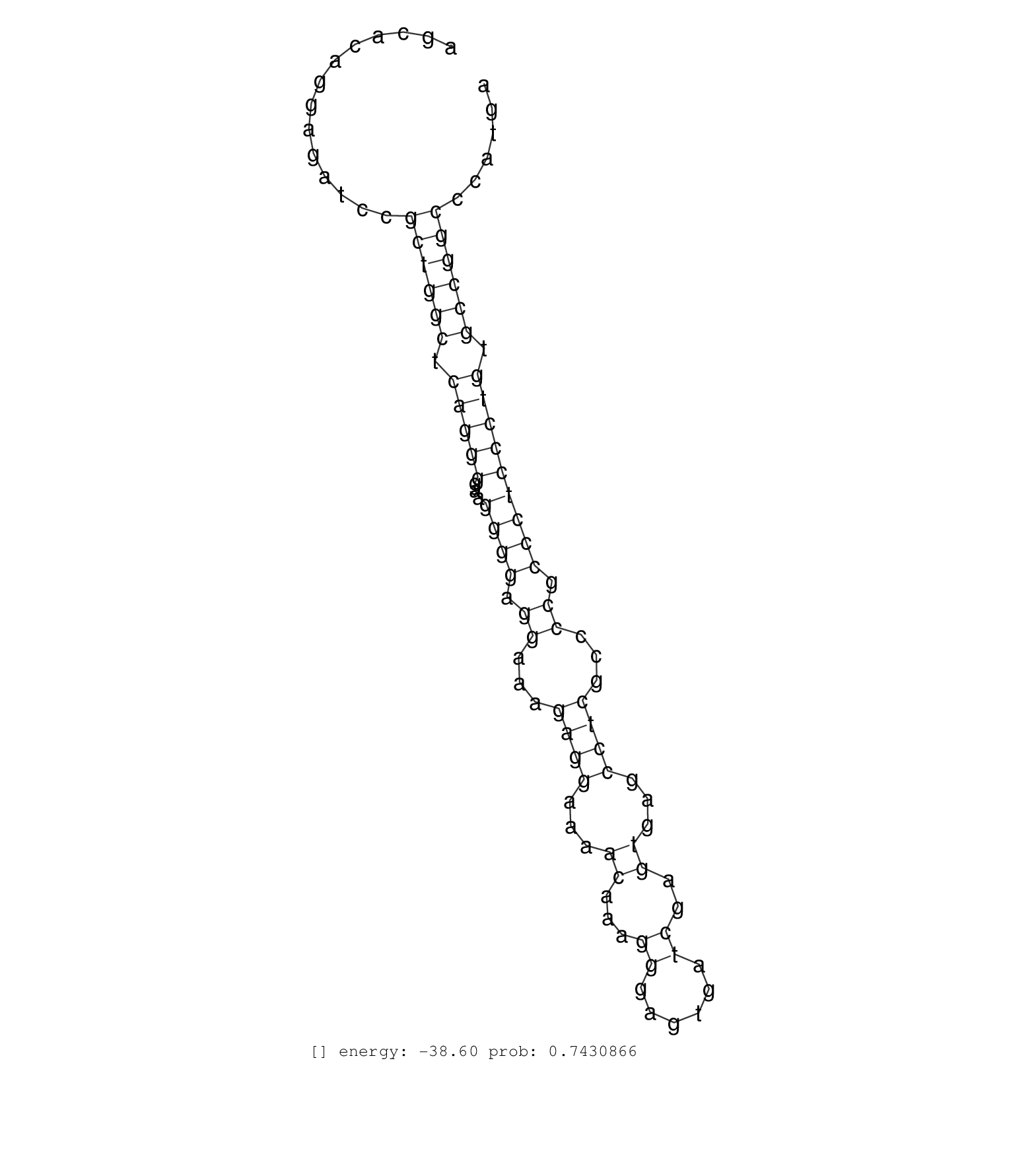
| AGTGGGCTTCAGTGCAGGGGCTTTTGCACTGGGCTGCAAGCCCTGAGCAGGTGACCTGAGCAGGTGTGTGACTAGTGCCTGTCTGTCTGCAATAGCCCTGACCCTCTAATCTGCATCCTGTGAGCCACCAAGACTGAACCAAGCCCAGGAAGCGGCGCCACCAACCGGCCTCGCAGAGGCAGAGGCCCTGGAGCAGTGACAGCACAGGAGATCCGCTGGCTCAGGGGAAGGGGAGGAAAGAGGAAAACAAAGGGAGTGATCGAGTGAGCCTCGCCCCGCCCTCCCTGTGCCGGCCCATGATGTGTCAGTCAGAGGCGCGGCGAGGACCAGAGCTCCGTGCAGCGAAATGG ......................................................................................................................................................................................................................((((((.(((((...((((.((...((((...((...((......))..))...))))...)).))))))))).))))))........................................................ ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553607(SRX182813) source: Testis. (testes) | SRR553581(SRX182787) source: Testis. (testes) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553580(SRX182786) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................CACCAAGACTGAACCAAGCCCAGGAAGCGGCGC................................................................................................................................................................................................ | 33 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - |
| ..........................................................................................................TAATCTGCATCCTGTGAGCCACCAAGAC........................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..............................................................................................................................ACCAAGACTGAACCAAGCCCAGGAAGCGG................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ..............................................................................................................................ACCAAGACTGAACCAAGCCCAGGAAGCGGCGC................................................................................................................................................................................................ | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ......................................................................................................................................TGAACCAAGCCCAGGAAGCGGCGCCACCAA.......................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ......................................................................................................................................TGAACCAAGCCCAGGAAGCGGCGCCA.............................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ......................................................................................................................TGTGAGCCACCAAGACTGAACCAAGCCCAGACGC...................................................................................................................................................................................................... | 34 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| ..................................................................................................................................................................................................................................................................................................................................AGGACCAGAGCTCCGTGC.......... | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - |
| ......................................................................................................................................TGAACCAAGCCCAGGAAGCGGCGCCAC............................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ...................................................................................................................................GACTGAACCAAGCCCAGGAAGCGGC.................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ......................................................................................................................TGTGAGCCACCAAGACTGAACCAAGC.............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ................................................................................................................................CAAGACTGAACCAAGCCCAGGAAGCGGCGCC............................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| AGTGGGCTTCAGTGCAGGGGCTTTTGCACTGGGCTGCAAGCCCTGAGCAGGTGACCTGAGCAGGTGTGTGACTAGTGCCTGTCTGTCTGCAATAGCCCTGACCCTCTAATCTGCATCCTGTGAGCCACCAAGACTGAACCAAGCCCAGGAAGCGGCGCCACCAACCGGCCTCGCAGAGGCAGAGGCCCTGGAGCAGTGACAGCACAGGAGATCCGCTGGCTCAGGGGAAGGGGAGGAAAGAGGAAAACAAAGGGAGTGATCGAGTGAGCCTCGCCCCGCCCTCCCTGTGCCGGCCCATGATGTGTCAGTCAGAGGCGCGGCGAGGACCAGAGCTCCGTGCAGCGAAATGG ......................................................................................................................................................................................................................((((((.(((((...((((.((...((((...((...((......))..))...))))...)).))))))))).))))))........................................................ ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553607(SRX182813) source: Testis. (testes) | SRR553581(SRX182787) source: Testis. (testes) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553580(SRX182786) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................................tgaCGATCACTCCCTAGT..................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | 1.00 | |
| .................................................................................................................................................................................................ATCTCCTGTGCTGTCACTG.......................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - |
| .................................................................................................................................................................................................TCTCCTGTGCTGTCACTG........................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - |