| (1) HEART | (2) TESTES |
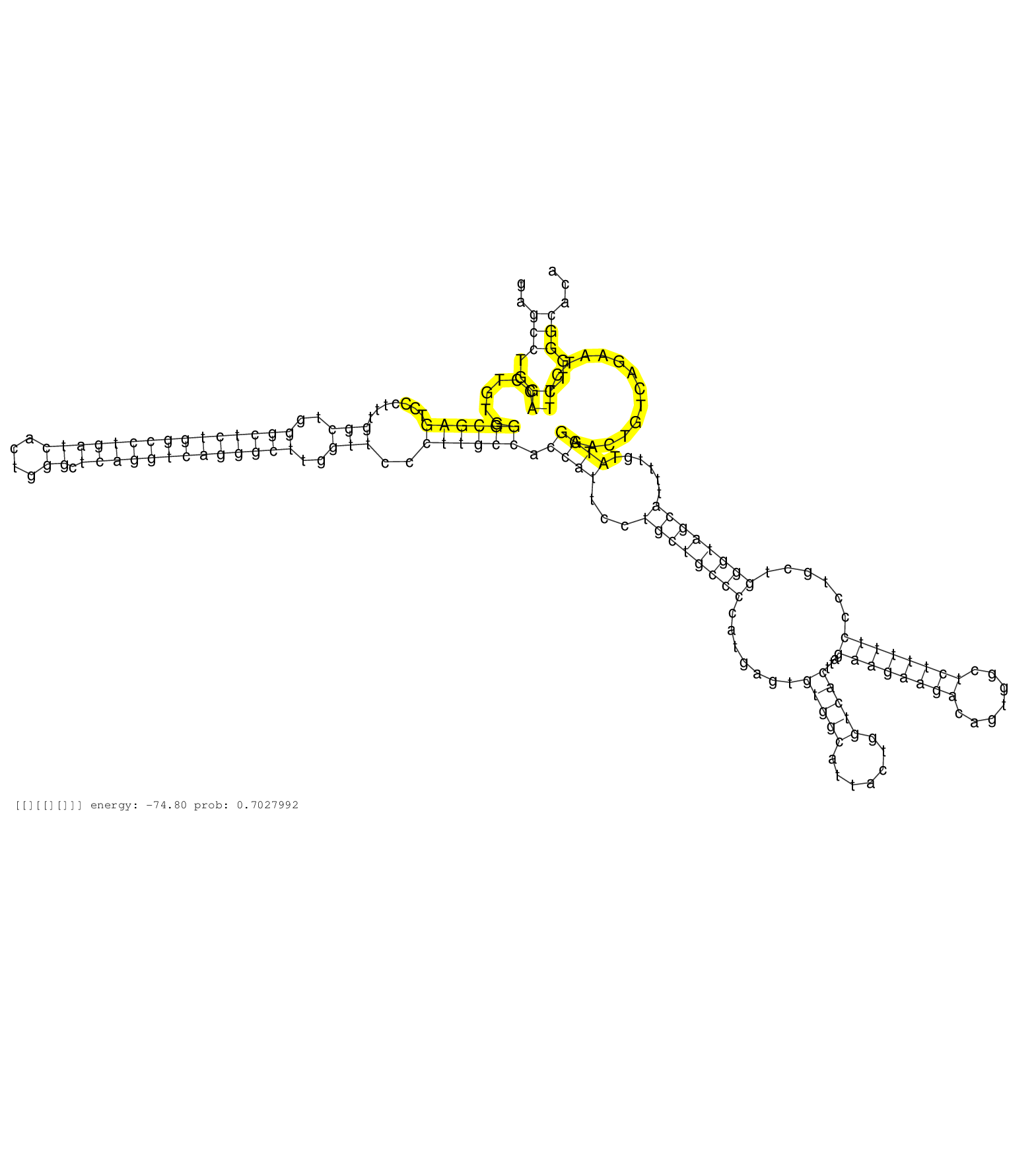
| GTGAATGAGGCTGTGCAGCTGGTCTTCCGCGAGGAGGGCTGGGCCTCGGAGTGAGGGCAGTGCGGGGGAGGCGAGGGGGTGAGCCTGGACCTGTGGGCGAGTCCCTTTGGCTGGGCTCTGGCCTGATCACTGGGCTCAGGTCAGGGCTTGGTTCCCTTGCCACCATTCCTGCTGCCCCATGAGTGTGGCATTACTGGTCACTTAGAAGAAGACAGTGGCTCTTTTTCCCTGCTGGGTAGCATTTTGTATGGAACTGTCAGAATCTTCTGGGCACAGTTCCCTTGTGCCTCTCCTGGCAGTCTAACCCCAGGCCATTCTCTTCACCTGTGTGCCTCAGTGTCCTTCTCGTT ..................................................................................(((..((......((((((.......(((..(((((((((((((((....)).)))))))))))))..)))..)))))).((((...((((((((.......(((((.......)))))...((((((((.......))))))))......))))))))......))))..............))..))).............................................................................. ................................................................................81................................................................................................................................................................................................275......................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553607(SRX182813) source: Testis. (testes) | SRR553581(SRX182787) source: Testis. (testes) | SRR553579(SRX182785) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................................TTTTGTATGGAACTGTCAGAATCTTCTGGGC.............................................................................. | 31 | 1 | 3.00 | 3.00 | 3.00 | - | - |
| ...................................................................................................................................................................................................................................................TTGTATGGAACTGTCAGAATCTTCTGGGC.............................................................................. | 29 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - |
| ..................................................................................................................................................................................................................................................TTTGTATGGAACTGTCAGAATCTTCTGGGC.............................................................................. | 30 | 1 | 2.00 | 2.00 | 2.00 | - | - |
| ............................................................................................................................................................................................................................................TAGCATTTTGTATGGAACTGTCAGAATCT..................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...........................................................................................................................TGATCACTGGGCTCAGGTCAGGGCTTG........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ......................................................................................................................................................................................................................................................TATGGAACTGTCAGAATCTTCTGGC............................................................................... | 25 | 1.00 | 0.00 | - | 1.00 | - | |
| .....................................................................................TGGACCTGTGGGCGAGTCC...................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| .......................................................................................................................................................................................................................................................ATGGAACTGTCAGAATCTTCTGGGC.............................................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ....................................................................................................................................................................................................................................................TGTATGGAACTGTCAGAATCTTCTGGGC.............................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| GTGAATGAGGCTGTGCAGCTGGTCTTCCGCGAGGAGGGCTGGGCCTCGGAGTGAGGGCAGTGCGGGGGAGGCGAGGGGGTGAGCCTGGACCTGTGGGCGAGTCCCTTTGGCTGGGCTCTGGCCTGATCACTGGGCTCAGGTCAGGGCTTGGTTCCCTTGCCACCATTCCTGCTGCCCCATGAGTGTGGCATTACTGGTCACTTAGAAGAAGACAGTGGCTCTTTTTCCCTGCTGGGTAGCATTTTGTATGGAACTGTCAGAATCTTCTGGGCACAGTTCCCTTGTGCCTCTCCTGGCAGTCTAACCCCAGGCCATTCTCTTCACCTGTGTGCCTCAGTGTCCTTCTCGTT ..................................................................................(((..((......((((((.......(((..(((((((((((((((....)).)))))))))))))..)))..)))))).((((...((((((((.......(((((.......)))))...((((((((.......))))))))......))))))))......))))..............))..))).............................................................................. ................................................................................81................................................................................................................................................................................................275......................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553607(SRX182813) source: Testis. (testes) | SRR553581(SRX182787) source: Testis. (testes) | SRR553579(SRX182785) source: Heart. (Heart) |
|---|