| (1) KIDNEY | (2) OTHER | (2) TESTES |
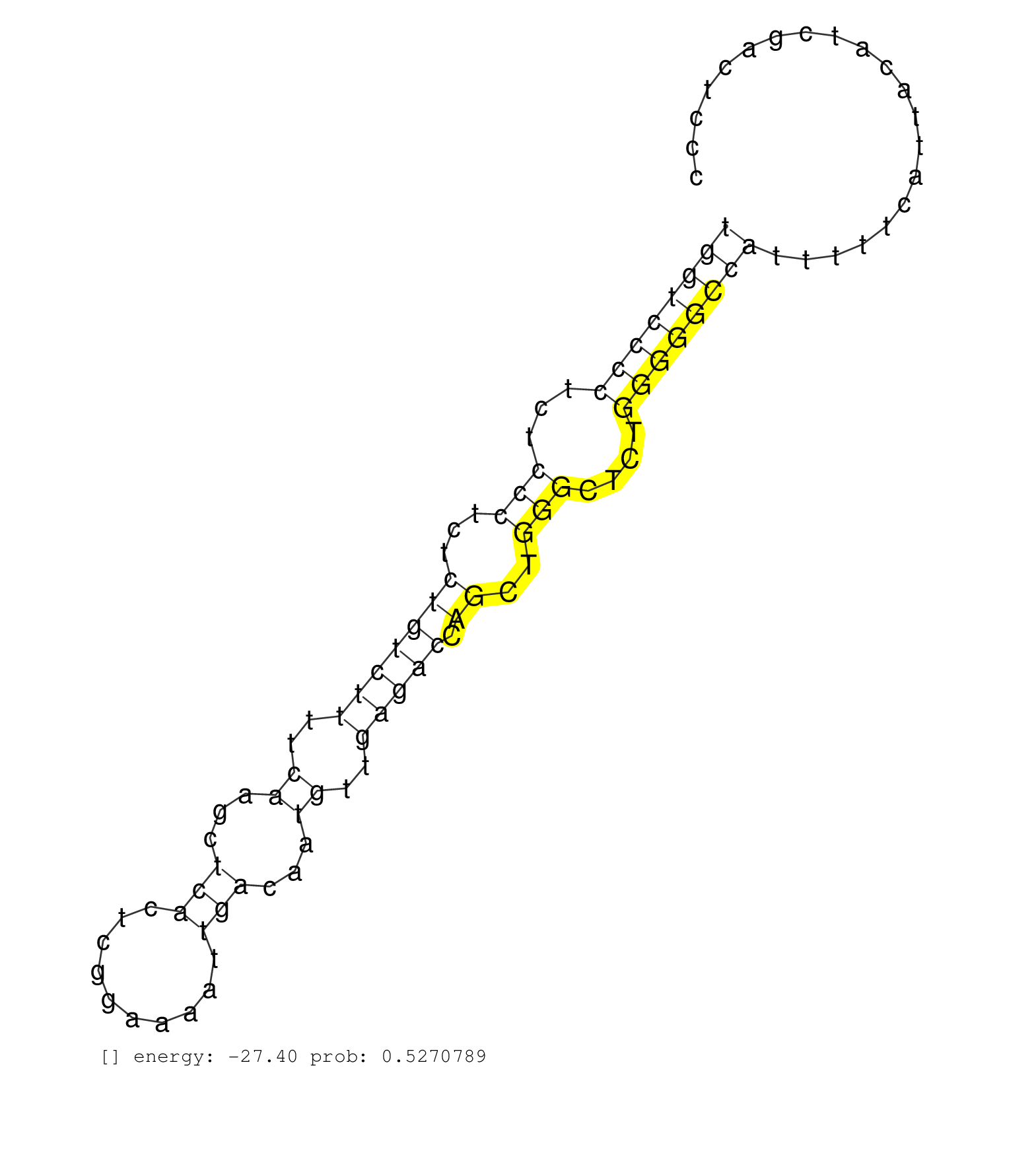
| TCAGCTTGCTACTCCAGGAGTTCGGCTTTCCCAGGAGCAAAGTGCTGAAGGTAAGGGTGCCCTCAGGGCACACCCCAGCCCCTTTATGGAGTGAGGGGCCTGAAACCACTTCTCTAGTTACTGGGGTTGTGTCCCCCAGCCCATCCCTGGCTTCCAAGGAGCCGCAGCCTCTGATGCACAAGCCTTTCCTGTCTGCGTGATGGTCCCCTCTCCCTCTCTGTCTTTTCAAGCTCACTCGGAAAATTGACAATGTTGAGACCAGCTGGGCTCTGGGGGCCATTTTTCATTACATCGACTCCCTGAACAGACAGAAGAGTCCAGCCTCATAGCAGCCGAGCCACCCCTGTCCC ........................................................................................................................................................................................................((((((((...(((...(((((((..((...(((..........)))...))..))))).))..)))....))))))))....................................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553607(SRX182813) source: Testis. (testes) | SRR553581(SRX182787) source: Testis. (testes) | SRR553577(SRX182783) source: Frontal Cortex. (Frontal Cortex) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) | SRR553580(SRX182786) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................................................TCGGAAAATTGACAATGTTGAGACCAGC....................................................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................TACATCGACTCCCTGAACAGACAGAAGA................................... | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - |
| ...............................................................................................................................................................................................................................................................................................TACATCGACTCCCTGAACAGACAGAAGAGT................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................ACATCGACTCCCTGAACAG........................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - |
| ..........................................................................................................................................................................................................................................CTCGGAAAATTGACAATGTTGAGACC.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 |
| ........................................................................................................................................................................................................................................CACTCGGAAAATTGACAATGTTGAGACCAGC....................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - |
| ..........................................................................................................................................................................................................................................CTCGGAAAATTGACAATGTTGAGACCAGC....................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ...................................................................................................................................................................................................................................................................CAGCTGGGCTCTGGGGGC......................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - |
| TCAGCTTGCTACTCCAGGAGTTCGGCTTTCCCAGGAGCAAAGTGCTGAAGGTAAGGGTGCCCTCAGGGCACACCCCAGCCCCTTTATGGAGTGAGGGGCCTGAAACCACTTCTCTAGTTACTGGGGTTGTGTCCCCCAGCCCATCCCTGGCTTCCAAGGAGCCGCAGCCTCTGATGCACAAGCCTTTCCTGTCTGCGTGATGGTCCCCTCTCCCTCTCTGTCTTTTCAAGCTCACTCGGAAAATTGACAATGTTGAGACCAGCTGGGCTCTGGGGGCCATTTTTCATTACATCGACTCCCTGAACAGACAGAAGAGTCCAGCCTCATAGCAGCCGAGCCACCCCTGTCCC ........................................................................................................................................................................................................((((((((...(((...(((((((..((...(((..........)))...))..))))).))..)))....))))))))....................................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553607(SRX182813) source: Testis. (testes) | SRR553581(SRX182787) source: Testis. (testes) | SRR553577(SRX182783) source: Frontal Cortex. (Frontal Cortex) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) | SRR553580(SRX182786) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|---|
| .......TGGGAAAGCCGAACTCCTGGAGTAGC............................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |