| (2) KIDNEY | (1) TESTES |
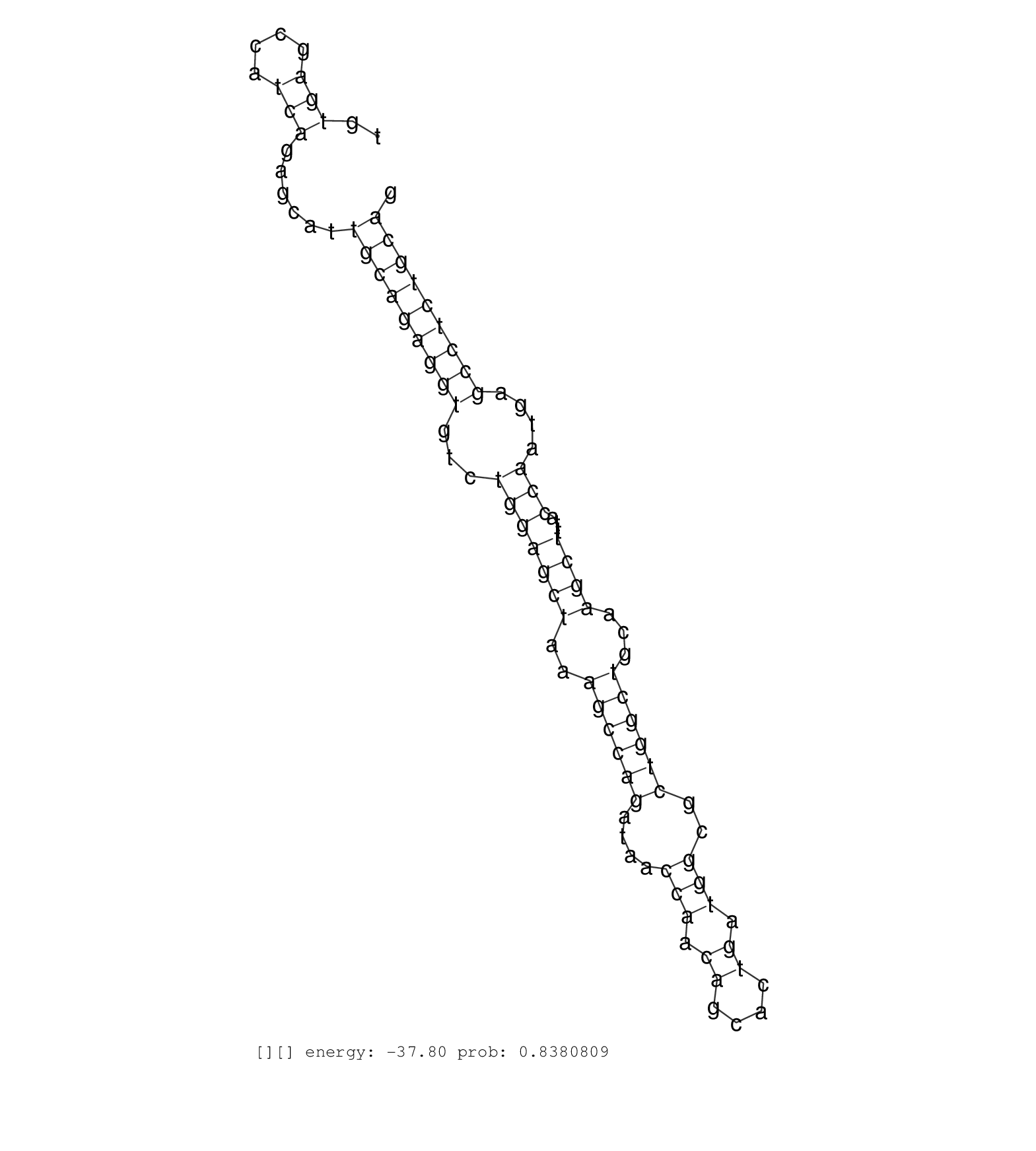
| GTACTCTCACTTTATTTCTCAGATTGGCCGACACTTAGGGAAAATAGAAAAGTACTCATGTTGAATTATCAGGGACAGGTTCCCCCGATAGTCCCTGCCAAATGCTGTGCTGCTTTTTGAGGCAGCTATGCAGCAGAATCCTAAGAATATGGAAGCTTGGCAGTACCTGGGTACCACCCAGGCAGAGAATGAACAAGAACTGTGAGCCATCAGAGCATTGCAGAGGTGTCTGGAGCTAAAGCCAGATAACCAACAGCACTGATGGCGCTGGCTGCAAGCTTTACCAATGAGCCTCTGCAGCAGCAGGCCTGTGAAACCCTGCTACACACCAGCCTGTGCCTATCTGGTGA ..........................................................................................................................................................................................................(((....)))......(((((((((...(((((((..((((((....(((.((....)).)))..))))))...))))...)))....)))))))))................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553580(SRX182786) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................TGAACAAGAACTGTGAGCCATCAGAGC...................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 4.00 | - | - |
| ......................................................................................................................................................................................CAGAGAATGAACAAGAACTGTGAGCC.............................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - |
| ........................................................................................................................................................................................GAGAATGAACAAGAACTGTGAGCCATC........................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ....................................AGGGAAAATAGAAAAGTACTCATGTT................................................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .........................................................................................................................................................................................AGAATGAACAAGAACTGTGAGCCATC........................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .............................................................................................................................................................................................TGAACAAGAACTGTGAGCCATCAGAGCATT................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .........................................................................................................................................................................................................................................................................................TACCAATGAGCCTCTGCAGCAGCAGGC.......................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .......................................................................................................................................................................................AGAGAATGAACAAGAACTGTGAGCCATC........................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ....................................................................................................................................................................................GGCAGAGAATGAACAAGAACTGTGAGCCATC........................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..............................................GAAAAGTACTCATGTAGG.............................................................................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | 1.00 | - | |
| .............................................................................................................................................................................................TGAACAAGAACTGTGAGCCATCAGAGCAT.................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ................................................................................................................................................................................................ACAAGAACTGTGAGC............................................................................................................................................... | 15 | 12 | 0.08 | 0.08 | - | - | 0.08 |
| GTACTCTCACTTTATTTCTCAGATTGGCCGACACTTAGGGAAAATAGAAAAGTACTCATGTTGAATTATCAGGGACAGGTTCCCCCGATAGTCCCTGCCAAATGCTGTGCTGCTTTTTGAGGCAGCTATGCAGCAGAATCCTAAGAATATGGAAGCTTGGCAGTACCTGGGTACCACCCAGGCAGAGAATGAACAAGAACTGTGAGCCATCAGAGCATTGCAGAGGTGTCTGGAGCTAAAGCCAGATAACCAACAGCACTGATGGCGCTGGCTGCAAGCTTTACCAATGAGCCTCTGCAGCAGCAGGCCTGTGAAACCCTGCTACACACCAGCCTGTGCCTATCTGGTGA ..........................................................................................................................................................................................................(((....)))......(((((((((...(((((((..((((((....(((.((....)).)))..))))))...))))...)))....)))))))))................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553580(SRX182786) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................................................................................................CAGGCCTGCTGCTGCAG....................................... | 17 | 6 | 0.17 | 0.17 | 0.17 | - | - |