| (2) KIDNEY | (2) TESTES |
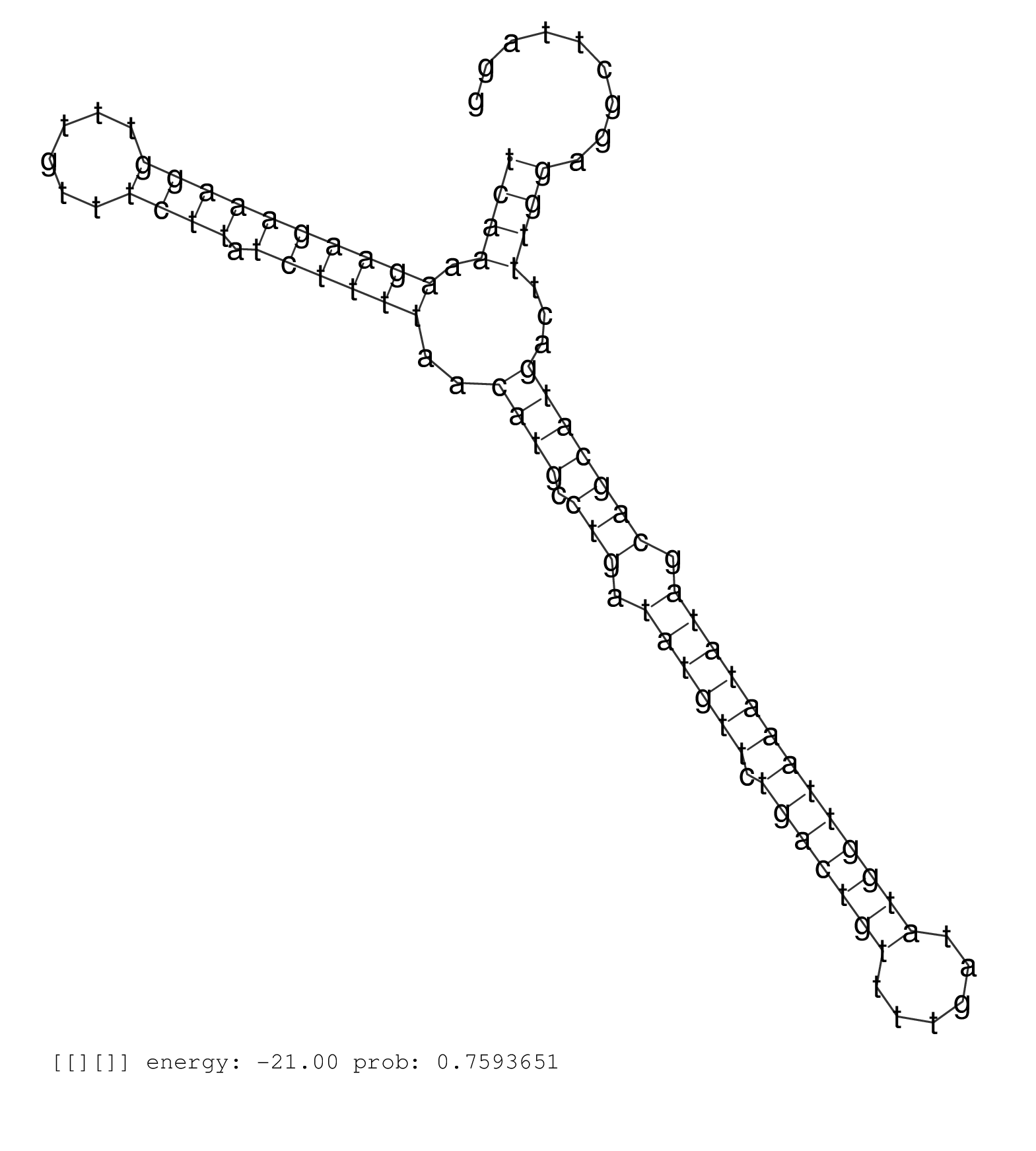
| AAATTTTAAGGCTCCTTTTCAAACATCAGGGGAAAATGGGAAAGGCTGTCGTGACTCAAAGACCCCATCTGAGTCCATTGTGGCCATTTCTGAGTGTCACACCTTACTTTCTTGTAAGGTGCAGCTGTTGGGGAGCCAAGAAAGTGAATGTCCAGACTCGGTACTGCGTGACGTTCTATCTGGAGGAAGACAGACATATGTCAAAAGAAGAAAGGTTTGTTTCTTATCTTTTAACATGCCTGATATGTTCTGACTGTTTTGATATGGTTAAATATAGCAGCATGACTTTGGAGGCTTAGGAGTGCATGTGTTTTTGAGGAAGACCTCTAAATTCGTATACTTGCATTGAG ........................................................................................................................................................................................................((((.((((((((((......)))).))))))..((((.(((.((((((.(((((((......))))))))))))).)))))))...))))........................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553580(SRX182786) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................TGCGTGACGTTCTATCTGGAGGAAGACAGT............................................................................................................................................................ | 30 | 2.00 | 0.00 | 2.00 | - | - | - | |
| ....................................TGGGAAAGGCTGTCGTGACTCAAAGACCC............................................................................................................................................................................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - |
| ...........................................................................................................................................................ACTCGGTACTGCGTGACG................................................................................................................................................................................. | 18 | 1 | 2.00 | 2.00 | - | - | 2.00 | - |
| ...................................................................................................................................................................CTGCGTGACGTTCTATCTGGAGGAAG................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .................................................................................................................................................................TACTGCGTGACGTTCTATCTGGAGGAAGAG............................................................................................................................................................... | 30 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ............................................GCTGTCGTGACTCAAAGACCCCATCTGAGT.................................................................................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .............................................................................................................................................................TCGGTACTGCGTGACGTTCTATCT......................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ............................................GCTGTCGTGACTCAAAGAC............................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .....................................................................................................................................................................GCGTGACGTTCTATCTGGAGGAAGAC............................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...................................................................................................................................................................CTGCGTGACGTTCTATCTGGAGGAAGACA.............................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ....................................TGGGAAAGGCTGTCGTGACTCAAAGAC............................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..................................................................................................................................................AATGTCCAGACTCGGTACTGCGTGAC.................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .....................................................................................................................................................................GCGTGACGTTCTATCTGGAGGAAGACAGACA.......................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| AAATTTTAAGGCTCCTTTTCAAACATCAGGGGAAAATGGGAAAGGCTGTCGTGACTCAAAGACCCCATCTGAGTCCATTGTGGCCATTTCTGAGTGTCACACCTTACTTTCTTGTAAGGTGCAGCTGTTGGGGAGCCAAGAAAGTGAATGTCCAGACTCGGTACTGCGTGACGTTCTATCTGGAGGAAGACAGACATATGTCAAAAGAAGAAAGGTTTGTTTCTTATCTTTTAACATGCCTGATATGTTCTGACTGTTTTGATATGGTTAAATATAGCAGCATGACTTTGGAGGCTTAGGAGTGCATGTGTTTTTGAGGAAGACCTCTAAATTCGTATACTTGCATTGAG ........................................................................................................................................................................................................((((.((((((((((......)))).))))))..((((.(((.((((((.(((((((......))))))))))))).)))))))...))))........................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553580(SRX182786) source: Kidney. (Kidney) |
|---|