| (1) HEART | (2) KIDNEY | (2) OTHER |
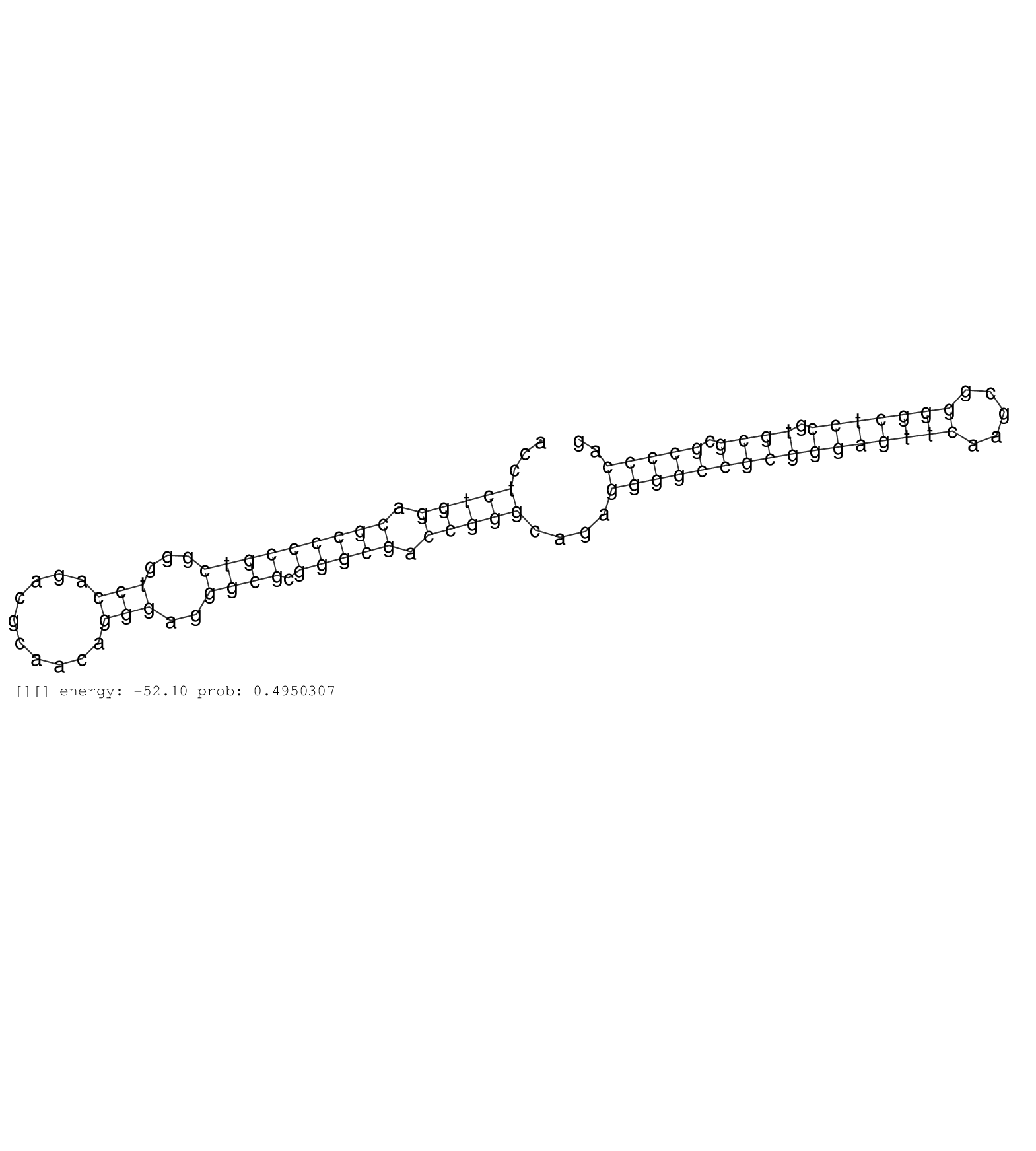
| TCACCCAGAGGACAGCCGGCGGGGAATTCCGAGGGTGGGAGTGAGGAGAGGTAGGAGAGGCCAGGGCAGAGCGAGGCCCCGCGTAGGATAGGAACCATCGCCCGGTGCGAGCCTGACCTTCCAGGGCCGGCAACTCCTCGGCAGAGCGACAGCGCGGGGTCTCAGAGCGCGGCTCGGAGCCGCACTAGGAGCGCTGGACCGTGGGGGAGAAGCTCGGAGGGCGGGTAGCTCCCAGTAAAGCGGCCTAGCGGGTAGGTACAGGGCCCCTCCCCTCGCTAGTCCTCATTGCCCGTTCTCCCTGCGCCGGGGCGCTCCAGGCCCCAGCCCTAACCCGGGCCCTAGCGGCTCCCCACTCCATGACCTCTGGACGCCCCGTCGGGTCCAGACGCAACAGGGAGGGCGCGGGCGACCGGGCAGAGGGGCCGCGGGAGTTCAAGCGGGGCTCCGTGCGCGCCCCAGCCGCCCGCCTGTGCTGTCCCTGCAGATGTCGGGAGCTATCTTCGGGCCCC ..........................................................................................................................................................................................................................................................................................................................................................................(((((.(((((((((...(((..........)))..)))).))))).)))))....((((((((((((((((.....))))))).)))).))))).................................................... .......................................................................................................................................................................................................................................................................................................................................................................360................................................................................................459................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) | SRR553579(SRX182785) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|---|
| ................................................................................GCGTAGGATAGGAACCAT........................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 5.00 | 5.00 | 2.00 | 2.00 | 1.00 | - | - |
| ...............................................................................CGCGTAGGATAGGAACCAT........................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 5.00 | 5.00 | 3.00 | 2.00 | - | - | - |
| ................................................................................GCGTAGGATAGGAAC.............................................................................................................................................................................................................................................................................................................................................................................................................................. | 15 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - |
| ...............................................................................CGCGTAGGATAGGAAC.............................................................................................................................................................................................................................................................................................................................................................................................................................. | 16 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| .............................................................................CCCGCGTAGGATAGGAACCAT........................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| .........................................................................AGGCCCCGCGTAGGA..................................................................................................................................................................................................................................................................................................................................................................................................................................... | 15 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - |
| .......................................................................................ATAGGAACCATCGCCCGG.................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..............................................................................................................................................................................CGGAGCCGCACTAGGAGCGCTGGGCC..................................................................................................................................................................................................................................................................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | 1.00 |
| TCACCCAGAGGACAGCCGGCGGGGAATTCCGAGGGTGGGAGTGAGGAGAGGTAGGAGAGGCCAGGGCAGAGCGAGGCCCCGCGTAGGATAGGAACCATCGCCCGGTGCGAGCCTGACCTTCCAGGGCCGGCAACTCCTCGGCAGAGCGACAGCGCGGGGTCTCAGAGCGCGGCTCGGAGCCGCACTAGGAGCGCTGGACCGTGGGGGAGAAGCTCGGAGGGCGGGTAGCTCCCAGTAAAGCGGCCTAGCGGGTAGGTACAGGGCCCCTCCCCTCGCTAGTCCTCATTGCCCGTTCTCCCTGCGCCGGGGCGCTCCAGGCCCCAGCCCTAACCCGGGCCCTAGCGGCTCCCCACTCCATGACCTCTGGACGCCCCGTCGGGTCCAGACGCAACAGGGAGGGCGCGGGCGACCGGGCAGAGGGGCCGCGGGAGTTCAAGCGGGGCTCCGTGCGCGCCCCAGCCGCCCGCCTGTGCTGTCCCTGCAGATGTCGGGAGCTATCTTCGGGCCCC ..........................................................................................................................................................................................................................................................................................................................................................................(((((.(((((((((...(((..........)))..)))).))))).)))))....((((((((((((((((.....))))))).)))).))))).................................................... .......................................................................................................................................................................................................................................................................................................................................................................360................................................................................................459................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) | SRR553579(SRX182785) source: Heart. (Heart) |
|---|