| (1) HEART | (1) OTHER |
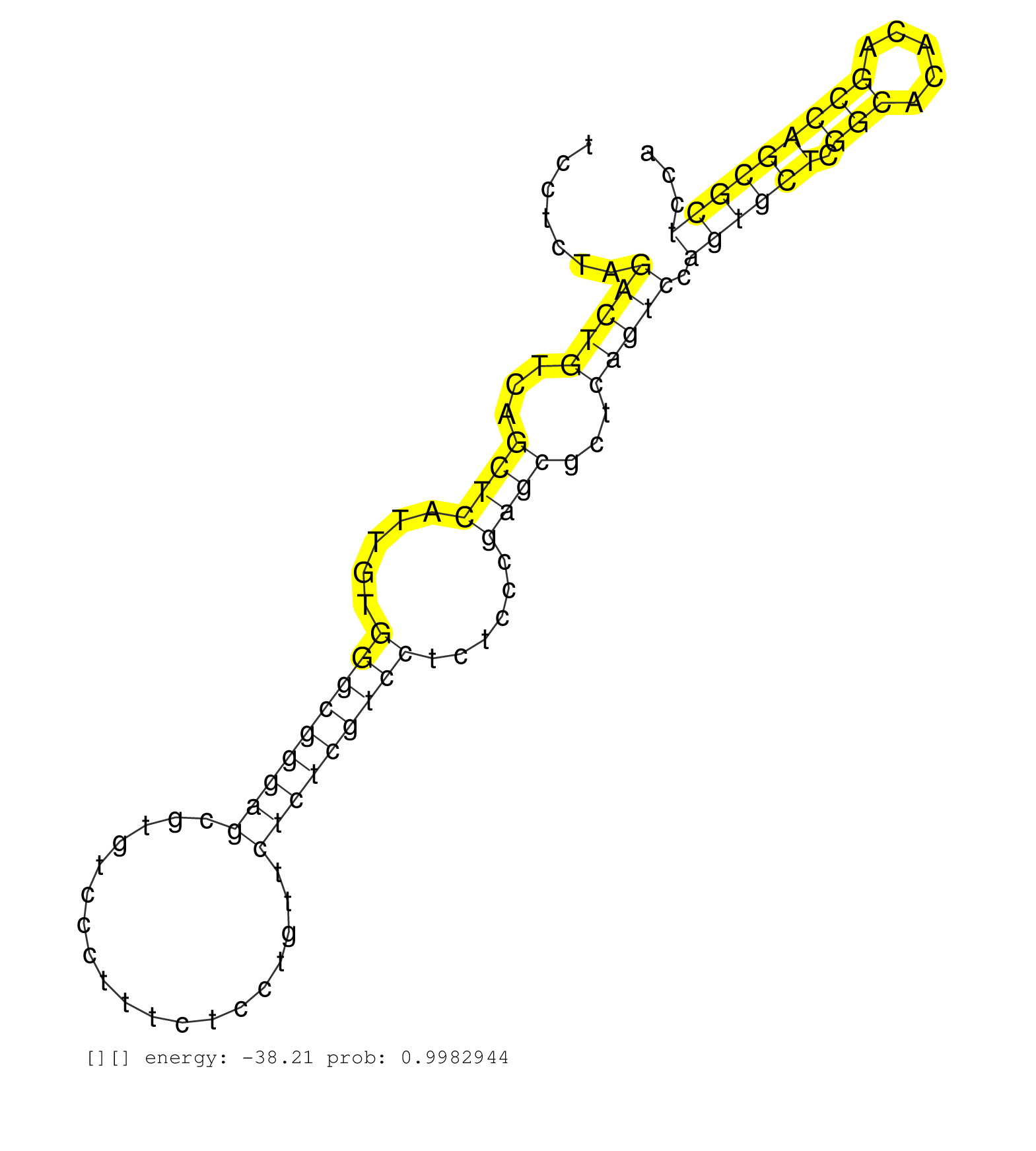
| NNNNNNNNNNCAGGCCCGGGGATCTAGCCGGCTAGGCACTCCTCTCAACCCTCTCTGAAGGACCCATCGACGCCTCAGCGGGGCCGACGGGTTTCACGCTTCCTCTAGACTGTCAGCTCATTGTGGGCGGGAGCGTGTCCCTTTCTCCTGTTCTCTCGTCCTCTCCCGAGCGCTCAGTCCAGTGCTCGGCACACAGCCAGCGCTCCACAGACACGATCGACCGAGCGAGTGACGAGGTCGTGTCGCCGCGCGTGCGGCCGCGAGCCCGGCCGGTGACGCGGCCGTTTCCTCTCCCCGCAGCGCACGTCCGTCCTGGATTCCAGCGCCCTGAAGACGCGAGTCCAGCTGAG ...........................................................................................................(((((...((((.....(((((((((...................)))))))))......))))...))))).((((((.(((.....))))))))).................................................................................................................................................. ....................................................................................................101.......................................................................................................207............................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|
| .........................................................................................................TAGACTGTCAGCTCATTGGGC................................................................................................................................................................................................................................ | 21 | 5.00 | 0.00 | 5.00 | - | |
| .........................................................................................................TAGACTGTCAGCTCATTGTGCGCA............................................................................................................................................................................................................................. | 24 | 2.00 | 0.00 | 2.00 | - | |
| .........................................................................................................TAGACTGTCAGCTCATTGGGCA............................................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | 1.00 | - | |
| .........................................................................................................TAGACTGTCAGCTCATTGTGCA............................................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | 1.00 | - | |
| .........................................................................................................TAGACTGTCAGCTCATTGTTGGT.............................................................................................................................................................................................................................. | 23 | 1.00 | 0.00 | 1.00 | - | |
| .........................................................................................................TAGACTGTCAGCTCATTGTGCGCC............................................................................................................................................................................................................................. | 24 | 1.00 | 0.00 | 1.00 | - | |
| .........................................................................................................TAGACTGTCAGCTCATTGCGGC............................................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | 1.00 | - | |
| .........................................................................................................................................................................................................................................................................................................................TGGATTCCAGCGCCCTGAAGACGCCAG.......... | 27 | 1.00 | 0.00 | 1.00 | - | |
| .........................................................................................................TAGACTGTCAGCTCATTGTGGGGGGA........................................................................................................................................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | |
| ........................................................................................................................................................................................CTCGGCACACAGCCAGCGC................................................................................................................................................... | 19 | 2 | 0.50 | 0.50 | 0.50 | - |
| ..............................................................................................................................................................................................................................................................................................................CACGTCCGTCCTGGA................................. | 15 | 2 | 0.50 | 0.50 | - | 0.50 |
| .........................................................................................................TAGACTGTCAGCTCATTGTGGGCG............................................................................................................................................................................................................................. | 24 | 10 | 0.10 | 0.10 | 0.10 | - |
| ...........................................................................................................GACTGTCAGCTCATTGTGGGCGG............................................................................................................................................................................................................................ | 23 | 15 | 0.07 | 0.07 | - | 0.07 |
| NNNNNNNNNNCAGGCCCGGGGATCTAGCCGGCTAGGCACTCCTCTCAACCCTCTCTGAAGGACCCATCGACGCCTCAGCGGGGCCGACGGGTTTCACGCTTCCTCTAGACTGTCAGCTCATTGTGGGCGGGAGCGTGTCCCTTTCTCCTGTTCTCTCGTCCTCTCCCGAGCGCTCAGTCCAGTGCTCGGCACACAGCCAGCGCTCCACAGACACGATCGACCGAGCGAGTGACGAGGTCGTGTCGCCGCGCGTGCGGCCGCGAGCCCGGCCGGTGACGCGGCCGTTTCCTCTCCCCGCAGCGCACGTCCGTCCTGGATTCCAGCGCCCTGAAGACGCGAGTCCAGCTGAG ...........................................................................................................(((((...((((.....(((((((((...................)))))))))......))))...))))).((((((.(((.....))))))))).................................................................................................................................................. ....................................................................................................101.......................................................................................................207............................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|