| (1) OTHER |
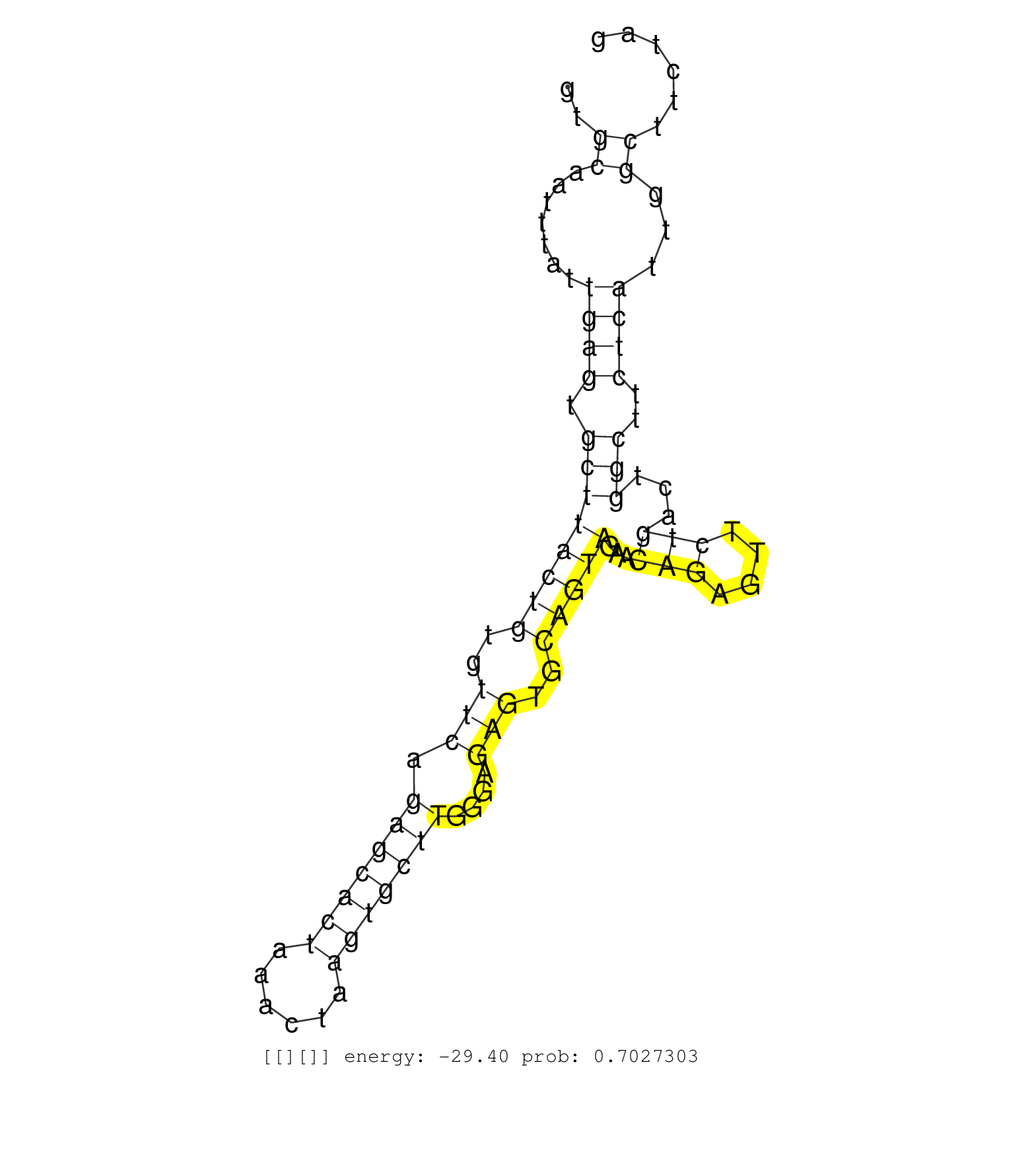
| AAATGCTCACCTGACTGACTGACCGACCGCCCAACCCTTCTGACTGCTTGACTGACTGCCTGGCCAACTGCCCGCCCACCTGACTGCCCAACTGAGTGCTTGACTGCCCACCTGACCGTCCAAACGCCTGACAACTTGACCCACCTGAATGAGGCATCTCTGGCTCTGCCCCATCGAGGGCCCCTTCAATCAATCTATCAGTGCAATTTATTGAGTGCTTACTGTGTTCAGAGCACTAAACTAAGTGCTTGGGAGAGTGCAGTAGAACAGAGTTCTGACTGGCTTCTCATTGGCTTCTAGGATCTACGACAACTTGCCCGAGGTGAGAAAGAAAAAAGAAGAGCAAATGA ..........................................................................................................................................................................................................((.......((((.((((((((..(((.(((((((......)))))))....)))..)))))...(((....)))...)))..))))...))........................................................ ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|
| .........................................................................................................................................................................................................................................................TGGGAGAGTGCAGTAGAACAGAGTTCTGA........................................................................ | 29 | 1 | 14.00 | 14.00 | 14.00 |
| .........................................................................................................................................................................................................................................................TGGGAGAGTGCAGTAGAACAGAGTTCTGACT...................................................................... | 31 | 1 | 5.00 | 5.00 | 5.00 |
| .........................................................................................................................................................................................................................................................TGGGAGAGTGCAGTAGAACAGAGTTCTGAC....................................................................... | 30 | 1 | 4.00 | 4.00 | 4.00 |
| .........................................................................................................................................................................................................................................................TGGGAGAGTGCAGTAGAACAGAGTTCTGACA...................................................................... | 31 | 2.00 | 4.00 | 2.00 | |
| .........................................................................................................................................................................................................................................................TGGGAGAGTGCAGTAGAACAGAGTTCTG......................................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 |
| .........................................................................................................................................................................................................................................................TGGGAGAGTGCAGTAGAACAGAGTTCA.......................................................................... | 27 | 1.00 | 1.00 | 1.00 | |
| .........................................................................................................................................................................................................................................................TGGGAGAGTGCAGTAGAACAGAGTTC........................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 |
| ..........................................................................................................................................................................................................................................................GGGAGAGTGCAGTAGAACAGAGTTCTGAC....................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 |
| .........................................................................................................................................................................................................................................................TGGGAGAGTGCAGTAGAACAGAGTTCTGT........................................................................ | 29 | 1.00 | 2.00 | 1.00 | |
| .........................................................................................................................................................................................................................................................TGGGAGAGTGCAGTAGAACAGAGTT............................................................................ | 25 | 8 | 0.12 | 0.12 | 0.12 |
| AAATGCTCACCTGACTGACTGACCGACCGCCCAACCCTTCTGACTGCTTGACTGACTGCCTGGCCAACTGCCCGCCCACCTGACTGCCCAACTGAGTGCTTGACTGCCCACCTGACCGTCCAAACGCCTGACAACTTGACCCACCTGAATGAGGCATCTCTGGCTCTGCCCCATCGAGGGCCCCTTCAATCAATCTATCAGTGCAATTTATTGAGTGCTTACTGTGTTCAGAGCACTAAACTAAGTGCTTGGGAGAGTGCAGTAGAACAGAGTTCTGACTGGCTTCTCATTGGCTTCTAGGATCTACGACAACTTGCCCGAGGTGAGAAAGAAAAAAGAAGAGCAAATGA ..........................................................................................................................................................................................................((.......((((.((((((((..(((.(((((((......)))))))....)))..)))))...(((....)))...)))..))))...))........................................................ ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) |
|---|