| (1) HEART | (1) KIDNEY | (2) OTHER |
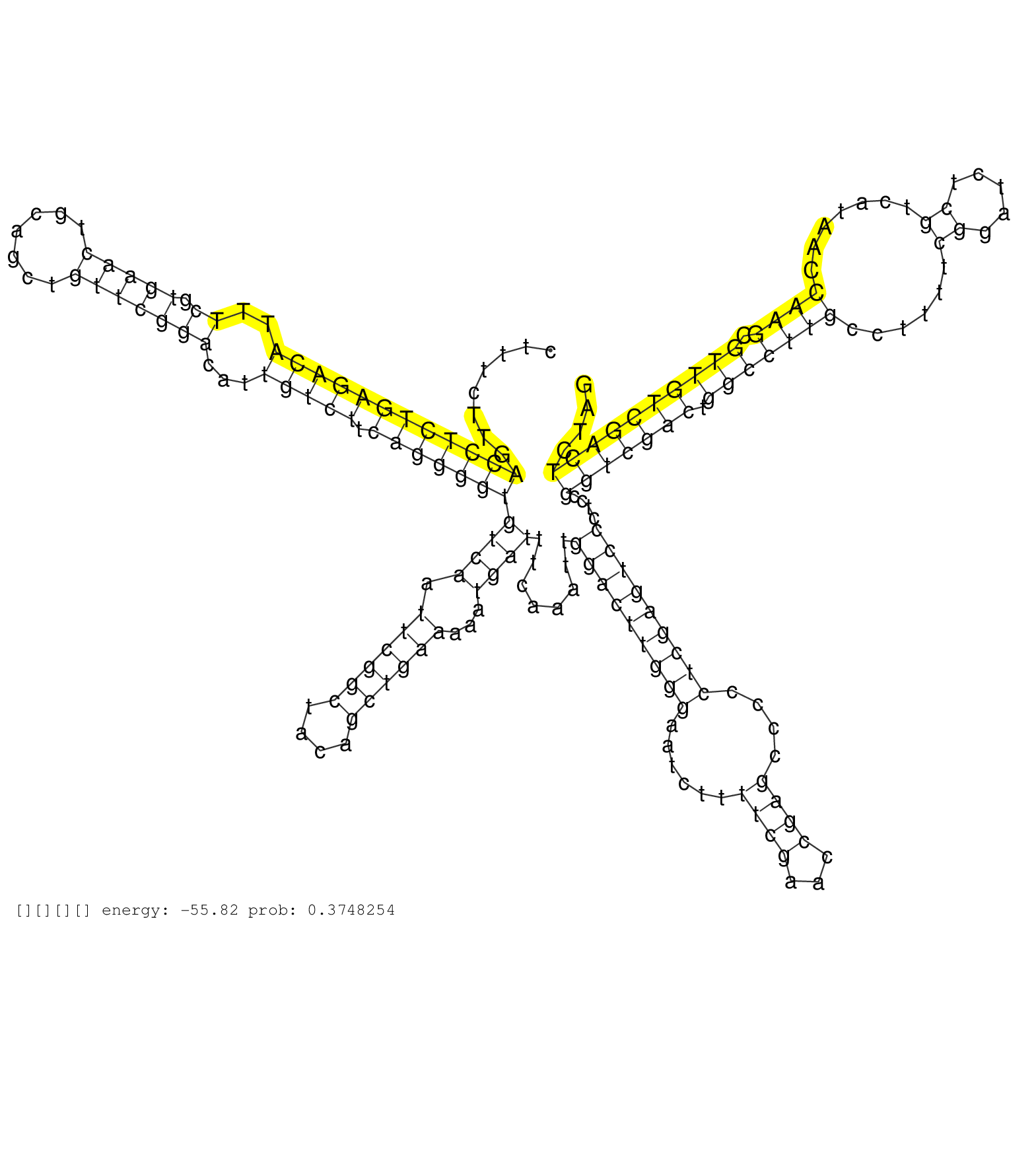
| TCTCCCGAGCGCTTAGTCCAGTGCCCTGCACGCGGTAAGCGCGCGATAAACGCCTCCGATCGATGGATGTTTTGAAATCGTATGTCATTGGACGCCCGATTCACCCTTTCTTGACCTCTGAGACATTTCGTGAACTGCAGCTGTTCGGACATTGTCTTCAGGGGTGTCAATTCGGCTACAGCTGAAAAATGATTTCAAATTGGACTTGGGAATCTTTTCGAACCGAGCCCCCTCGAGTCCCTCCGGTCGACTGGCCTTGCCTTTTCGGATCTCGTCATAACCAAGCGTTGTCGACTCTAGGTATCTGTCCGAGTTTCTTTATGCCTGGCTGATGTCAACTTTGAGTCGTG .................................................................................................................((((((((((((..((.(((((.......)))))))...))))).)))))))((((.((((((....))))))...))))........(((((((((......((((...))))....)))))))))....(((((((.(((((((......((.....)).......)))).))))))))))...................................................... .........................................................................................................106...............................................................................................................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................TTGACCTCTGAGACAGTG.............................................................................................................................................................................................................................. | 18 | 4.00 | 0.00 | 4.00 | - | - | - | |
| ..............................................................................................................TTGACCTCTGAGACAGGC.............................................................................................................................................................................................................................. | 18 | 4.00 | 0.00 | 4.00 | - | - | - | |
| ..............................................................................................................TTGACCTCTGAGACAGGA.............................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ..............................................................................................................TTGACCTCTGAGACACGG.............................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ..............................................................................................................TTGACCTCTGAGACAGCG.............................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ......................................................................................................................................................................................................................................................................................AACCAAGCGTTGTCGACTCTAG.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ......................................................................................................................TGAGACATTTCGTGAACTGCAGCTGTTCGG.......................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ..............................................................................................................TTGACCTCTGAGACATGG.............................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ...............................................................................................................................TCGTGAACTGCAGCT................................................................................................................................................................................................................ | 15 | 3 | 0.67 | 0.67 | - | - | - | 0.67 |
| TCTCCCGAGCGCTTAGTCCAGTGCCCTGCACGCGGTAAGCGCGCGATAAACGCCTCCGATCGATGGATGTTTTGAAATCGTATGTCATTGGACGCCCGATTCACCCTTTCTTGACCTCTGAGACATTTCGTGAACTGCAGCTGTTCGGACATTGTCTTCAGGGGTGTCAATTCGGCTACAGCTGAAAAATGATTTCAAATTGGACTTGGGAATCTTTTCGAACCGAGCCCCCTCGAGTCCCTCCGGTCGACTGGCCTTGCCTTTTCGGATCTCGTCATAACCAAGCGTTGTCGACTCTAGGTATCTGTCCGAGTTTCTTTATGCCTGGCTGATGTCAACTTTGAGTCGTG .................................................................................................................((((((((((((..((.(((((.......)))))))...))))).)))))))((((.((((((....))))))...))))........(((((((((......((((...))))....)))))))))....(((((((.(((((((......((.....)).......)))).))))))))))...................................................... .........................................................................................................106...............................................................................................................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|
| .........aaTGCAGGGCACTGGACTAAGAA.............................................................................................................................................................................................................................................................................................................................. | 23 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ..........CGTGCAGGGCACTGGACTAAGC.............................................................................................................................................................................................................................................................................................................................. | 22 | 18 | 0.11 | 0.11 | - | - | 0.11 | - |