| (1) BRAIN | (1) HEART | (1) KIDNEY | (1) OTHER |
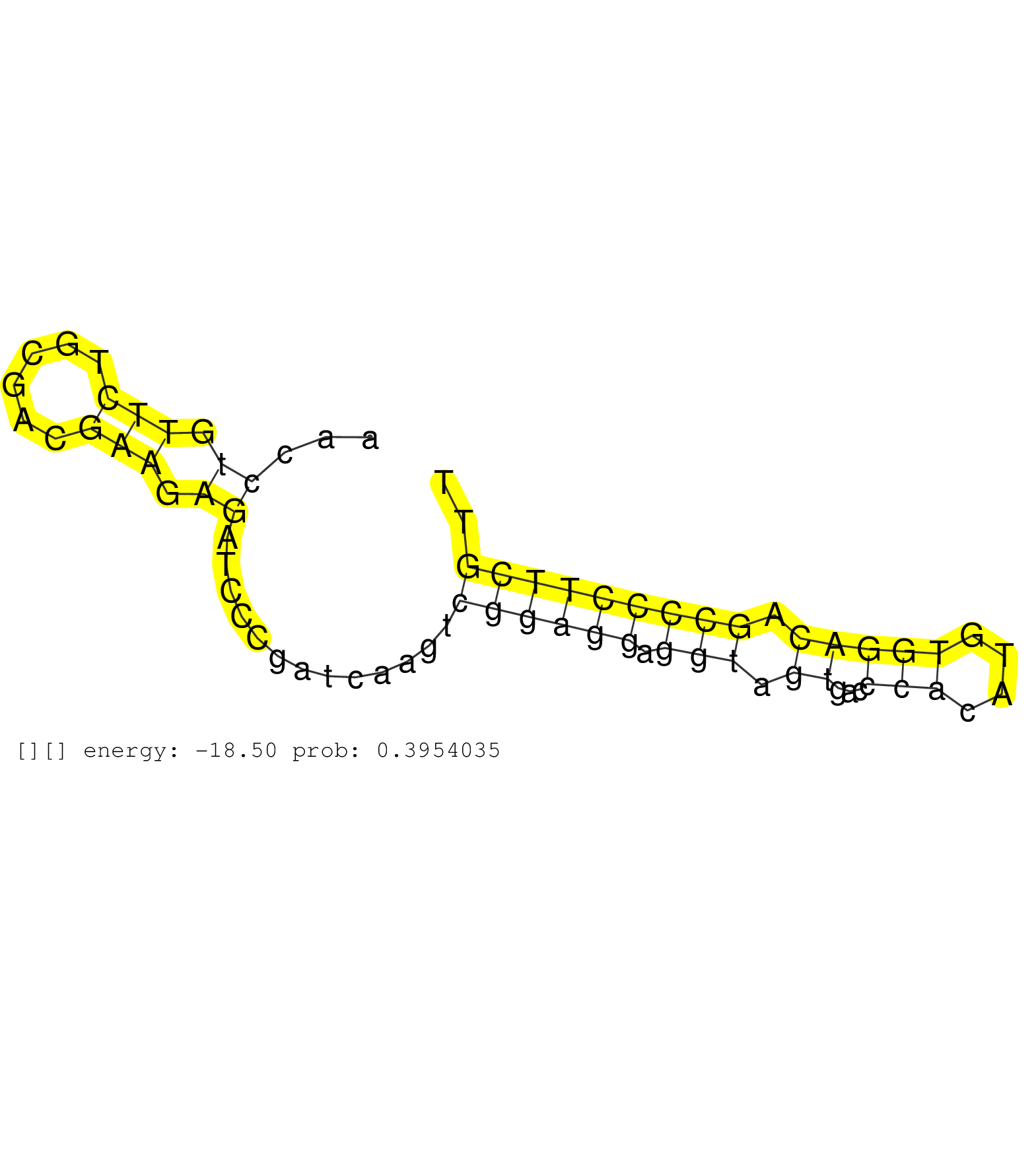
| GGTTTCCTCAGGACACTGCCAGCGTGAAATCCTCCAGCAGCCTGGAGCCGAACCTGTTCTGCGACGAAGAGATCCCGATCAAGTCGGAGGAGGTAGTGACCCACATGTGGACAGCCCCTTCGTTCTGTGCCGAACACGCGTATTCTTCGGCGTCCAAGAGCTGTTCCCAAGGTAAAGCCGCTTCCGGGGGTGGTGGGCGGGGGCAGAGACAGCGGAGGGCTCAGAGAGAGACGTTGTCGGTCGATCGGTGGCGTTTATTGCGTGCTCACCGTGTGCAGAGCACTGTACTAAGCGCTTGGGAGAGGACAGTACCGAGGAGTCGGTCCACACGTTCCCTGCCCAGAGGGAGG .....................................................((.(((......))).)).............((((((.(((.((...(((....))))).))))))))).................................................................................................................................................................................................................................... ..................................................51.......................................................................124................................................................................................................................................................................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553592(SRX182798) source: Brain. (Brain) |
|---|---|---|---|---|---|---|---|---|
| .......................................................GTTCTGCGACGAAGAGATCCC.................................................................................................................................................................................................................................................................................. | 21 | 1 | 2.00 | 2.00 | 2.00 | - | - | - |
| ...........................................................................CGATCAAGTCGGAGGAGGTAGTGAC.......................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ........................................................................................................ATGTGGACAGCCCCTTCGTTC................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ........................................................................................................................CGTTCTGTGCCGAACACGC................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .......................................................................ATCCCGATCAAGTCGGAGGAGGT................................................................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..................................................AACCTGTTCTGCGACGAAG......................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .............................................................................ATCAAGTCGGAGGAGGTAGTGACC......................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ..........................................TGGAGCCGAACCTGTTCTGCGACGAAGA........................................................................................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ...........................................................................................................TGGACAGCCCCTTCGTTCTGTGCCGAACACGC................................................................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| .............................................................................................TAGTGACCCACATGTGGACAGCCCCTTCGTTC................................................................................................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...........................................................................................................................................................................................................................................GTCGGTCGATCGGTGGGTC................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | 1.00 | |
| .......................................................GTTCTGCGACGAAGA........................................................................................................................................................................................................................................................................................ | 15 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .................................................................................................................................................................................................................................GAGAGACGTTGTCGG.............................................................................................................. | 15 | 2 | 0.50 | 0.50 | - | - | 0.50 | - |
| GGTTTCCTCAGGACACTGCCAGCGTGAAATCCTCCAGCAGCCTGGAGCCGAACCTGTTCTGCGACGAAGAGATCCCGATCAAGTCGGAGGAGGTAGTGACCCACATGTGGACAGCCCCTTCGTTCTGTGCCGAACACGCGTATTCTTCGGCGTCCAAGAGCTGTTCCCAAGGTAAAGCCGCTTCCGGGGGTGGTGGGCGGGGGCAGAGACAGCGGAGGGCTCAGAGAGAGACGTTGTCGGTCGATCGGTGGCGTTTATTGCGTGCTCACCGTGTGCAGAGCACTGTACTAAGCGCTTGGGAGAGGACAGTACCGAGGAGTCGGTCCACACGTTCCCTGCCCAGAGGGAGG .....................................................((.(((......))).)).............((((((.(((.((...(((....))))).))))))))).................................................................................................................................................................................................................................... ..................................................51.......................................................................124................................................................................................................................................................................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553592(SRX182798) source: Brain. (Brain) |
|---|