| (1) HEART | (1) KIDNEY | (1) OTHER |
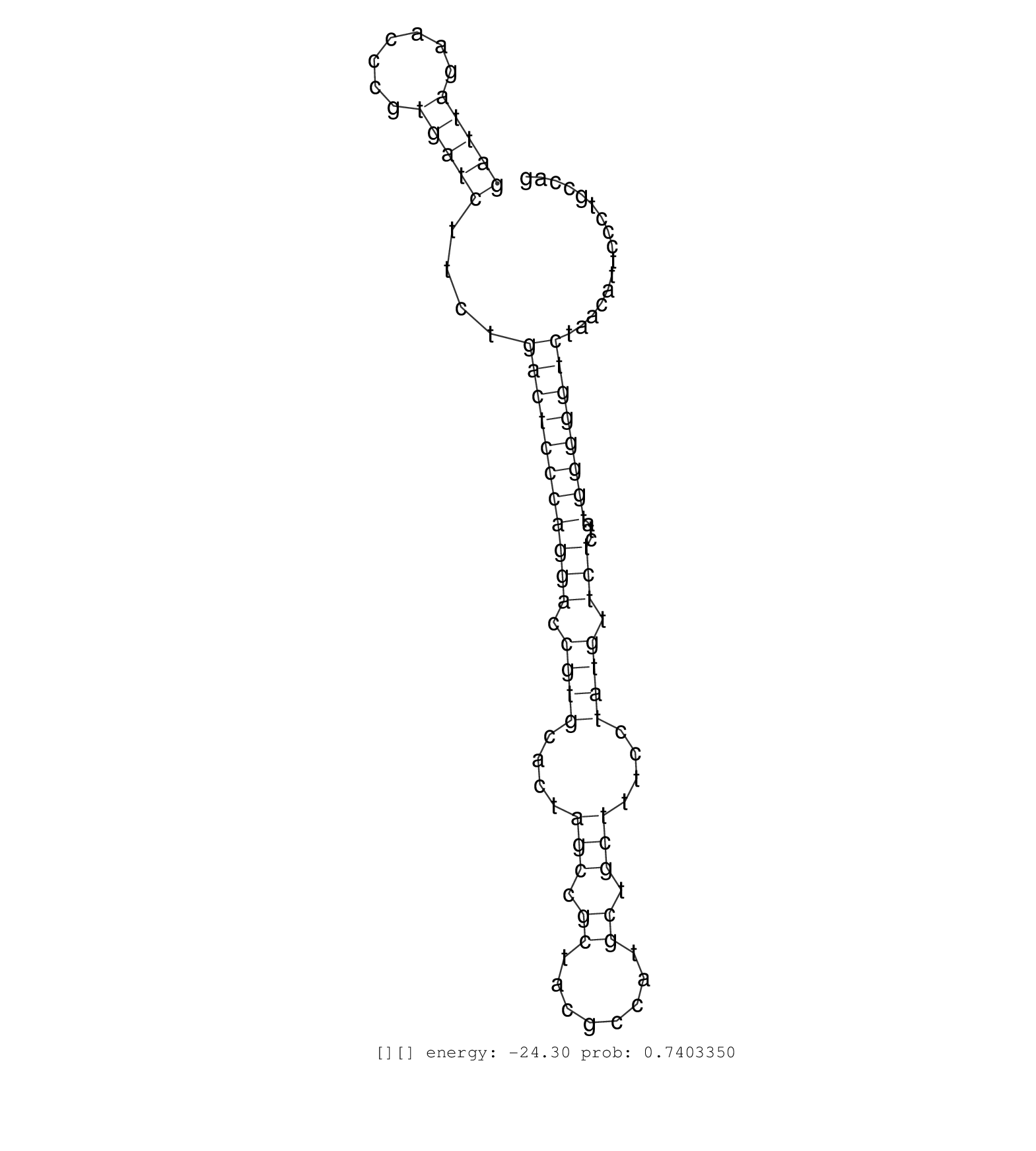
| GATTCGAACCCATGACCTCTGACTCCCAAGCCCCGGATCTTTCCGGCGCTGGGGTAGATACAAGCAAATCCGGTTGGACACAGTCCCTGTCCCACCTGGGGCTTACAGTCTGAATCCCCATTTTAAAGATGAGGTAACTGAGGCCCAGAGAAGTGAAGTGACTTGTCCAAGGTCACACAGCAGACACGTGGCGGAGCTGGGATTAGAACCCGTGATCTTCTGACTCCCAGGACCGTGCACTAGCCGCTACGCCATGCTGCTTTCCTATGTTCTCTATGGGGGTCTAACATTCCCTGCCAGGTACCGGATCGGCCCCTTCGAGGTGGAGAGTGCCCTGATCGAGCACCCGG ........................................................................................................................................................................................................(((((.......)))))....(((((((((((.((((....(((.((........)).)))....)))).)))...)))))))).................................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| ..........................................................TACAAGCAAATCCGGTTGGACACGGTA......................................................................................................................................................................................................................................................................... | 27 | 4.00 | 0.00 | 4.00 | - | - | |
| ..........................................................TACAAGCAAATCCGGTTGGACACGGA.......................................................................................................................................................................................................................................................................... | 26 | 3.00 | 0.00 | 3.00 | - | - | |
| ..........................................................TACAAGCAAATCCGGTTGGACACAATT......................................................................................................................................................................................................................................................................... | 27 | 2.00 | 0.00 | 2.00 | - | - | |
| .........................................................................................................................................................................AGGTCACACAGCAGACACGTTCAT............................................................................................................................................................. | 24 | 1.00 | 0.00 | - | - | 1.00 | |
| ..................................................................................................................................................................................................................................................................................................................................GTGGAGAGTGCCCTGATCGAG....... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ..........................................................TACAAGCAAATCCGGTTGGACACACTCG........................................................................................................................................................................................................................................................................ | 28 | 1.00 | 0.00 | 1.00 | - | - | |
| ..........................................................TACAAGCAAATCCGGTTGGACACTCGC......................................................................................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | 1.00 | - | - | |
| ..........................................................TACAAGCAAATCCGGTTGGACACGTCG......................................................................................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | 1.00 | - | - | |
| ............................................................................................................................................................................................TGGCGGAGCTGGGATTAGAACCGAGT........................................................................................................................................ | 26 | 1.00 | 0.00 | 1.00 | - | - | |
| .................................CGGATCTTTCCGGCGCTGGGGTAGATA.................................................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ..........................................................TACAAGCAAATCCGGTTGGACACGAAA......................................................................................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | 1.00 | - | - |
| GATTCGAACCCATGACCTCTGACTCCCAAGCCCCGGATCTTTCCGGCGCTGGGGTAGATACAAGCAAATCCGGTTGGACACAGTCCCTGTCCCACCTGGGGCTTACAGTCTGAATCCCCATTTTAAAGATGAGGTAACTGAGGCCCAGAGAAGTGAAGTGACTTGTCCAAGGTCACACAGCAGACACGTGGCGGAGCTGGGATTAGAACCCGTGATCTTCTGACTCCCAGGACCGTGCACTAGCCGCTACGCCATGCTGCTTTCCTATGTTCTCTATGGGGGTCTAACATTCCCTGCCAGGTACCGGATCGGCCCCTTCGAGGTGGAGAGTGCCCTGATCGAGCACCCGG ........................................................................................................................................................................................................(((((.......)))))....(((((((((((.((((....(((.((........)).)))....)))).)))...)))))))).................................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| ........................................................................................................TCATCTTTAAAATGGGGATTCAGACTGT.......................................................................................................................................................................................................................... | 28 | 10 | 0.10 | 0.10 | 0.10 | - | - |