| (1) HEART | (1) KIDNEY | (2) OTHER |
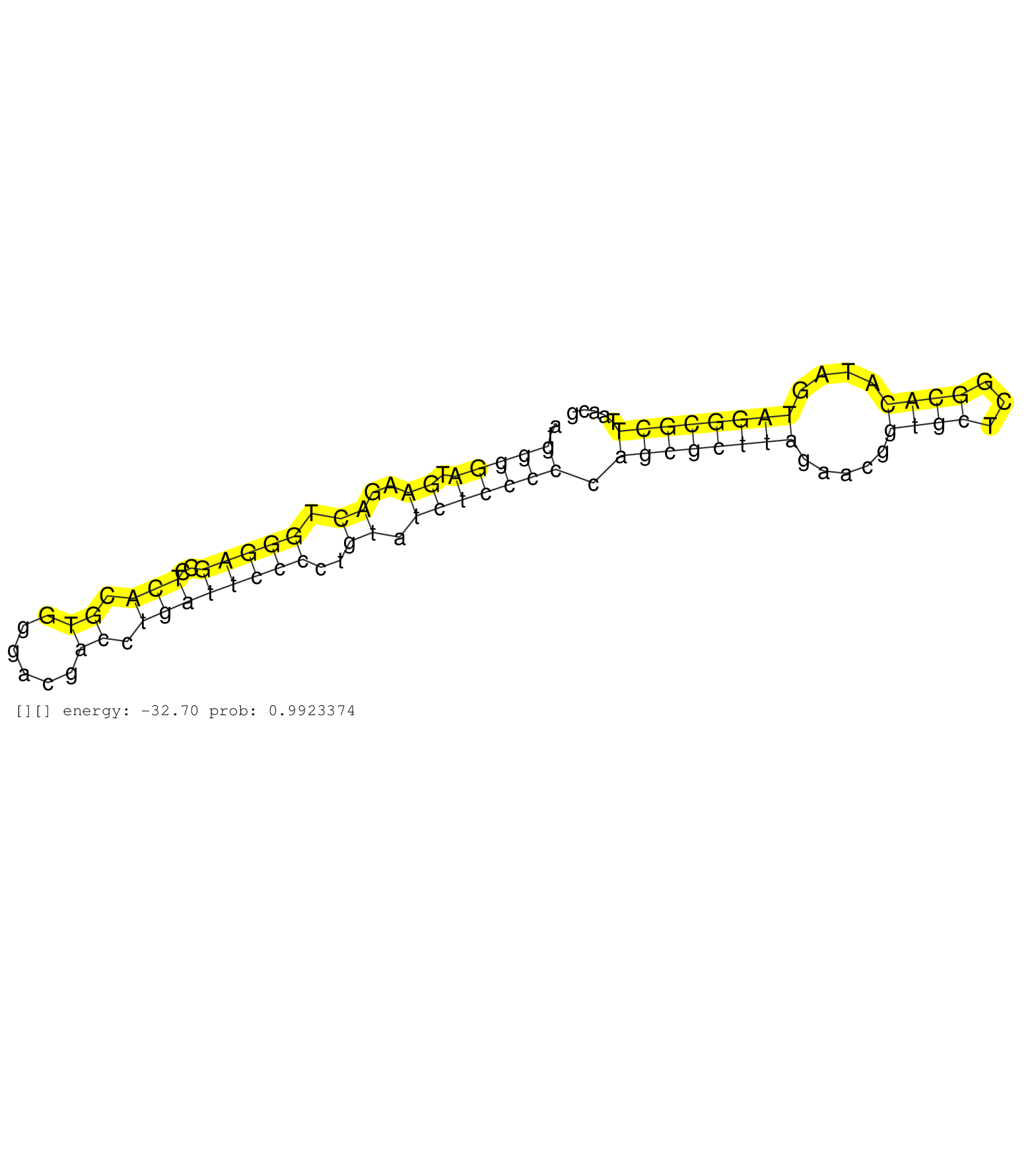
| TGGGGATGAAGACTGNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGTTCCCTCGTCTGGAAAATGGGGATGAAGACTGGGAGCCTCACGTGGGACGACCTGATTCCCCTGTATCTCCCCCAGCGCTTAGAACGGTGCTCGGCACATAGTAGGCGCTTAACGAATACCAACATTATTATTATTATTATTTTTATCCTTGAAGAATCTTCCAGAATCTAGGACTGACTTGACAGCACCGACTTTCCTGGCACCACCTCTATTTAAATCGCCTTCCTTTGCCCATTTTTCTTTGCTCTTTTAGGATTTTACGGTGTTTGGAGGCAGTCTGTCTGGAGCCCACGCCCAGAAGAT ................................................................(((((.((..((.(((((..(((.((......)).))))))))..)).))))))).((((((((.....((((...))))....)))))))).................................................................................................................................................................................................. ..............................................................63................................................................................................161........................................................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................................................................................................AGGATTTTACGGTGTTTG.................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ...................................................................GATGAAGACTGGGAGCCTCAATAA................................................................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ........................................................TGGAAAATGGGGATGAAGACTGGGTT............................................................................................................................................................................................................................................................................ | 26 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ...................................................................GATGAAGACTGGGAGCCTAAAA..................................................................................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ......................................................TCTGGAAAATGGGGATGAAGACTGGGAAAAA......................................................................................................................................................................................................................................................................... | 31 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .........................................................................................................................................TCGGCACATAGTAGGCGACA................................................................................................................................................................................................. | 20 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ......................................................TCTGGAAAATGGGGATGTGTA................................................................................................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ................................................................................................................................................................................................................................................................................................................................CAGTCTGTCTGGAGCCCACGC......... | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ......................................................TCTGGAAAATGGGGATGAAGAAAA................................................................................................................................................................................................................................................................................ | 24 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .................................................................GGGATGAAGACTGGGAGCCTGAAA..................................................................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ........................................................TGGAAAATGGGGATGAAGACTTGGT............................................................................................................................................................................................................................................................................. | 25 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .....................................................................TGAAGACTGGGAGCCTCACGTAAA................................................................................................................................................................................................................................................................. | 24 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ......................................................................GAAGACTGGGAGCCTCACGTAAA................................................................................................................................................................................................................................................................. | 23 | 1.00 | 0.00 | - | - | 1.00 | - | |
| ...........................................................................................................................................................................................................................................................................................................GGATTTTACGGTGTT.................................... | 15 | 4 | 0.25 | 0.25 | - | - | - | 0.25 |
| TGGGGATGAAGACTGNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGTTCCCTCGTCTGGAAAATGGGGATGAAGACTGGGAGCCTCACGTGGGACGACCTGATTCCCCTGTATCTCCCCCAGCGCTTAGAACGGTGCTCGGCACATAGTAGGCGCTTAACGAATACCAACATTATTATTATTATTATTTTTATCCTTGAAGAATCTTCCAGAATCTAGGACTGACTTGACAGCACCGACTTTCCTGGCACCACCTCTATTTAAATCGCCTTCCTTTGCCCATTTTTCTTTGCTCTTTTAGGATTTTACGGTGTTTGGAGGCAGTCTGTCTGGAGCCCACGCCCAGAAGAT ................................................................(((((.((..((.(((((..(((.((......)).))))))))..)).))))))).((((((((.....((((...))))....)))))))).................................................................................................................................................................................................. ..............................................................63................................................................................................161........................................................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|
| .....................................................................................tgtAGATACAGGGGAATCAGGTCGTCCCACTGT........................................................................................................................................................................................................................................ | 33 | 1.00 | 0.00 | - | 1.00 | - | - |