| (1) HEART | (1) KIDNEY |
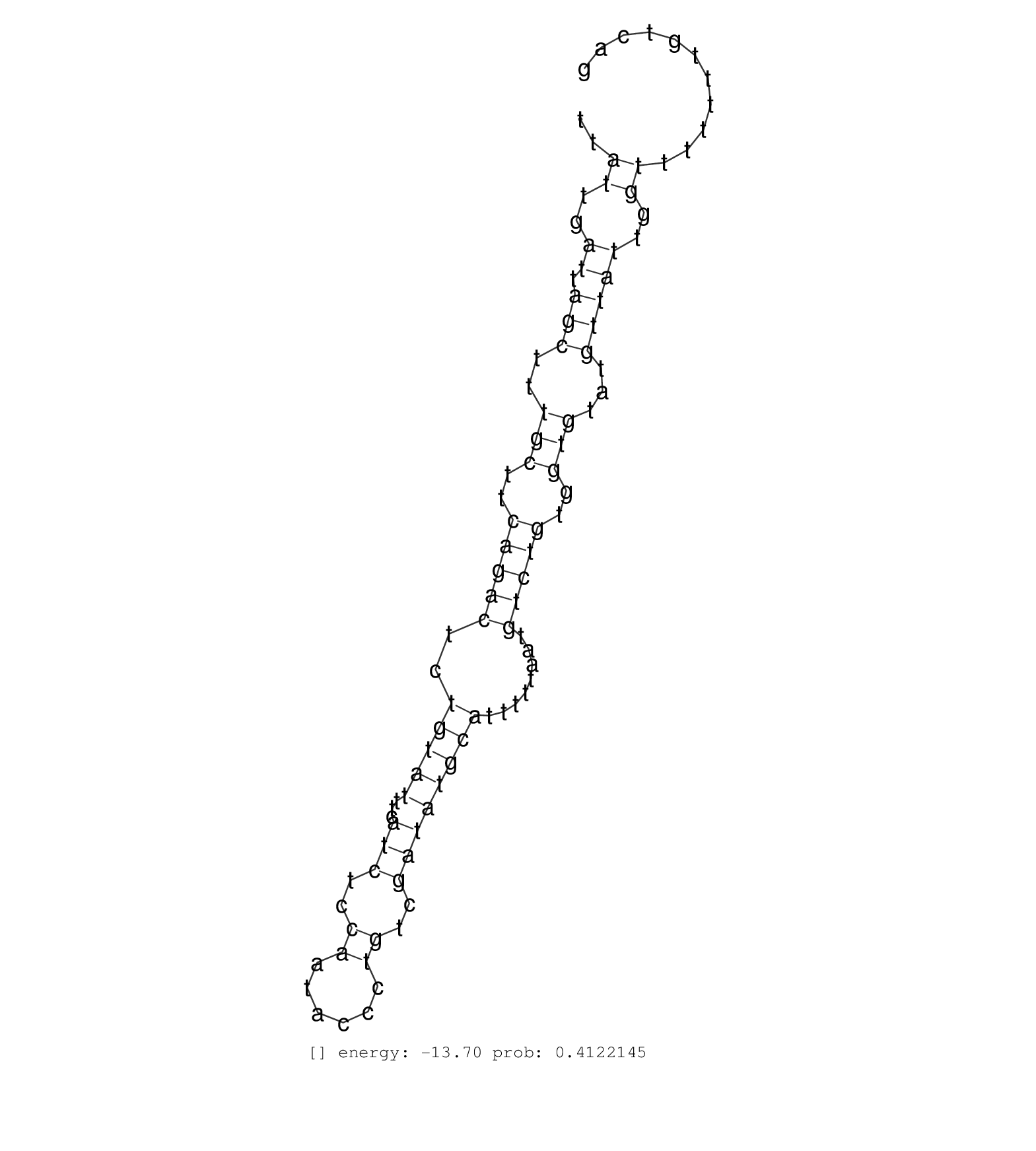
| GATATGCAGTTGCTAATAGTCTTCAGCCTCTCCCCAATCAGAATTTATGGGTGCGTTTTAGTCTCCAGACCAAGAACTCGGAAGTCGGTTACCCTAGATGTGCACAAAAGTGATGTGGCTTAATTTGGAAAGATGATAGAGTGGTTGGTGCTGTCAGCAGTACAGATGGAGAAGCTGGGATGAGGTGTTTTTTTTCTTTTTTATTGATTAGCTTTGCTTCAGACTCTGTATTTCATCTCCAATACCCTGTCGATATGCATTTTTAATGTCTGTGGTGTATGTTATTGGTTTTTTTGTCAGGTAATTCCTGCTCACTGTGTCTTTGCCAGTAACACATCTGCTCTCCCAAT ..........................................................................................................................................................................................................((..((.(((..(((..(((((..(((((...(((..((......))..))))))))........)))))..)))...)))))..))............................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|
| ..................................................................AGACCAAGAACTCGG............................................................................................................................................................................................................................................................................. | 15 | 2 | 1.00 | 1.00 | - | 1.00 |
| ..................................................................AGACCAAGAACTCGGG............................................................................................................................................................................................................................................................................ | 16 | 1.00 | 1.00 | 1.00 | - | |
| ....................................................................................................................................................................................................................................................................................................................TGCTCACTGTGTCTTTGCCAGTAACA................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - |
| .......................................................................................................................................................................................................................................................................................................................TCACTGTGTCTTTGCCAGTAACAC............... | 24 | 1 | 1.00 | 1.00 | 1.00 | - |
| .......................................................................AAGAACTCGGAAGTCGGTTACCCTAGATGT......................................................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - |
| .....................................................................................................................................................................................................................................................................................................................................CCAGTAACACATCTGCTCTCCC... | 22 | 1 | 1.00 | 1.00 | 1.00 | - |
| ..................................................................................................................................AGATGATAGAGTGGTTGGTGCTGTCAGCA............................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - |
| .......................................................................AAGAACTCGGAAGTCG....................................................................................................................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | 1.00 | - |
| .....................................................................................................................................................................................................................................................................................................................GCTCACTGTGTCTTTGCCAGTAACACATCT........... | 30 | 1 | 1.00 | 1.00 | 1.00 | - |
| ........................................................................................................................................................................................................................................................................................................................................GTAACACATCTGCTCTCC.... | 18 | 1 | 1.00 | 1.00 | 1.00 | - |
| ..............................................................................................................................................................................................................................................................................................................AATTCCTGCTCACTGTGTCTTTGCCAGTAACA................ | 32 | 1 | 1.00 | 1.00 | 1.00 | - |
| ..................................................................AGACCAAGAACTCGGAAG.......................................................................................................................................................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | 0.50 |
| ..................................................................AGACCAAGAACTCGGA............................................................................................................................................................................................................................................................................ | 16 | 2 | 0.50 | 0.50 | - | 0.50 |
| ...................................................................GACCAAGAACTCGGAAG.......................................................................................................................................................................................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | 0.50 |
| GATATGCAGTTGCTAATAGTCTTCAGCCTCTCCCCAATCAGAATTTATGGGTGCGTTTTAGTCTCCAGACCAAGAACTCGGAAGTCGGTTACCCTAGATGTGCACAAAAGTGATGTGGCTTAATTTGGAAAGATGATAGAGTGGTTGGTGCTGTCAGCAGTACAGATGGAGAAGCTGGGATGAGGTGTTTTTTTTCTTTTTTATTGATTAGCTTTGCTTCAGACTCTGTATTTCATCTCCAATACCCTGTCGATATGCATTTTTAATGTCTGTGGTGTATGTTATTGGTTTTTTTGTCAGGTAATTCCTGCTCACTGTGTCTTTGCCAGTAACACATCTGCTCTCCCAAT ..........................................................................................................................................................................................................((..((.(((..(((..(((((..(((((...(((..((......))..))))))))........)))))..)))...)))))..))............................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|