| (1) HEART | (1) KIDNEY | (2) OTHER |
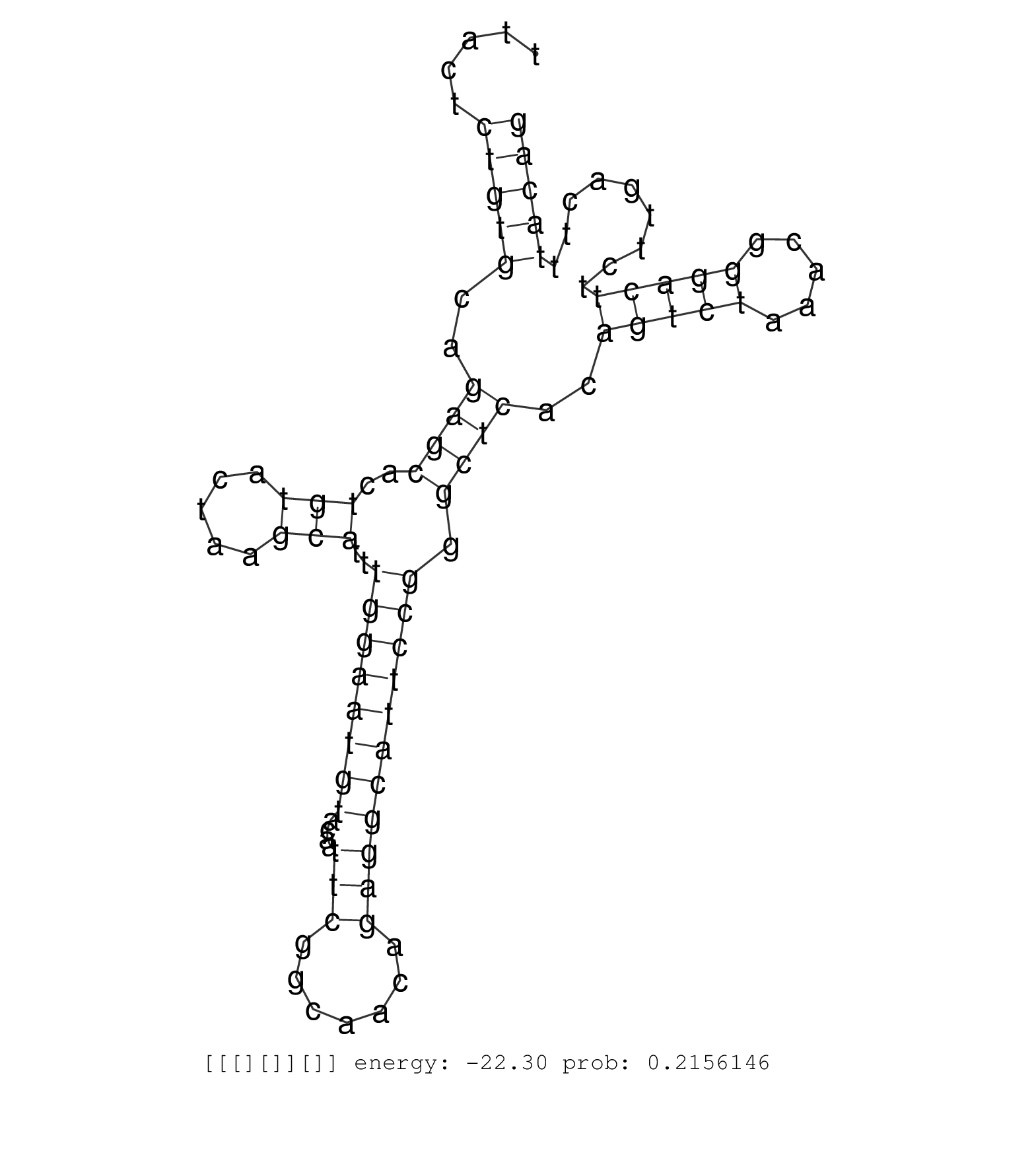
| GGAAACTGGAAATGTAAAGAAAATTAGATCACTTCCAGGAAGCTCTAAATAAGCTTAGAATTGTTTTTGACAAATTACCCAAACACTGGTTTAGTGGAGCTAGGATACCTGCTGTGTCTGTGATACCTGAACATGAGATACTAGACTGTGGTTCACTGCACATCAATTCTTTCATTCATTCAGTAGTATTTTTTGAGCGCTTACTCTGTGCAGAGCACTGTACTAAGCATTTGGAATGTACAATTCGGCAACAGAGGCATTCCGGGCTCACAGTCTAAACGGGACTTCTTGACTTTACAGGCAACAAATGGGATGTAATATACAGTGGAGCTGCTAGGGAACATCTCTGT .............................................................................................................................................................................................................(((((..((((..(((.....)))..((((((((....(((.......))))))))))).))))..(((((.....))))).........))))).................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................................TTGGAATGTACAATTCGGCAACAGAGGAG........................................................................................... | 29 | 3.00 | 0.00 | 3.00 | - | - | - | |
| .......................................................................................................................................................................................................................................TGGAATGTACAATTCGGCAACAGAGGAGA.......................................................................................... | 29 | 2.00 | 0.00 | 2.00 | - | - | - | |
| .......................................................................................................................................AGATACTAGACTGTGGTTCACTGCAC............................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| .................................................................................................................................................................................................TGAGCGCTTACTCTGTGCAGAGCAGAGA................................................................................................................................. | 28 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ......................................................................................................................................................................................................................................TTGGAATGTACAATTCGGCAACAGAGGAGAA......................................................................................... | 31 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ................................................................................................................................................................................................................................................................................................................CAAATGGGATGTAATATACAGTGGGCTG.................. | 28 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ........................................................................................................................TGATACCTGAACATGAGATACTAGACT........................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .......................................................................................................................................................................................................................................TGGAATGTACAATTCGGCAACAGAGGACA.......................................................................................... | 29 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ......................................................................................................................................................................................................................................TTGGAATGTACAATTCGGCAACAGAGGAGA.......................................................................................... | 30 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .....................................................................................................................................................................................................................................................................................................................GGGATGTAATATACAGTGGAGCTGCTAGG............ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ......................................................................................................................................................................................................................................TTGGAATGTACAATTCGGCAACAGAGTATA.......................................................................................... | 30 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ....................................................................................................................................................................................................................................................................................................................TGGGATGTAATATACAGTGGAGC................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ......................................................................................................................................................................................................................................TTGGAATGTACAATTCGGCAACAGAGTAAA.......................................................................................... | 30 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ....................................................................................................................................ATGAGATACTAGACTGTGGTTCACT................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ..................................................................................................................................................................................................................................................................................................................AATGGGATGTAATATACAGTGG...................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ................................................................................................................................GAACATGAGATACTAGACTGTGGTTCACTGCAC............................................................................................................................................................................................. | 33 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ......................................................................................................................................................................................................................................TTGGAATGTACAATTCGGCAACAGAGTATT.......................................................................................... | 30 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ..................................................................................................................................ACATGAGATACTAGACTGTGGTTCACTGC............................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ..................................................................................................................................................................................................................................................................................................................................CAGTGGAGCTGCTAGGGAACATC..... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| .................................................................................................................................................................................................................................................................................................................................ACAGTGGAGCTGCTAGG............ | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .............................................................................................GTGGAGCTAGGATACC................................................................................................................................................................................................................................................. | 16 | 3 | 0.33 | 0.33 | - | 0.33 | - | - |
| GGAAACTGGAAATGTAAAGAAAATTAGATCACTTCCAGGAAGCTCTAAATAAGCTTAGAATTGTTTTTGACAAATTACCCAAACACTGGTTTAGTGGAGCTAGGATACCTGCTGTGTCTGTGATACCTGAACATGAGATACTAGACTGTGGTTCACTGCACATCAATTCTTTCATTCATTCAGTAGTATTTTTTGAGCGCTTACTCTGTGCAGAGCACTGTACTAAGCATTTGGAATGTACAATTCGGCAACAGAGGCATTCCGGGCTCACAGTCTAAACGGGACTTCTTGACTTTACAGGCAACAAATGGGATGTAATATACAGTGGAGCTGCTAGGGAACATCTCTGT .............................................................................................................................................................................................................(((((..((((..(((.....)))..((((((((....(((.......))))))))))).))))..(((((.....))))).........))))).................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|