| (2) OTHER |
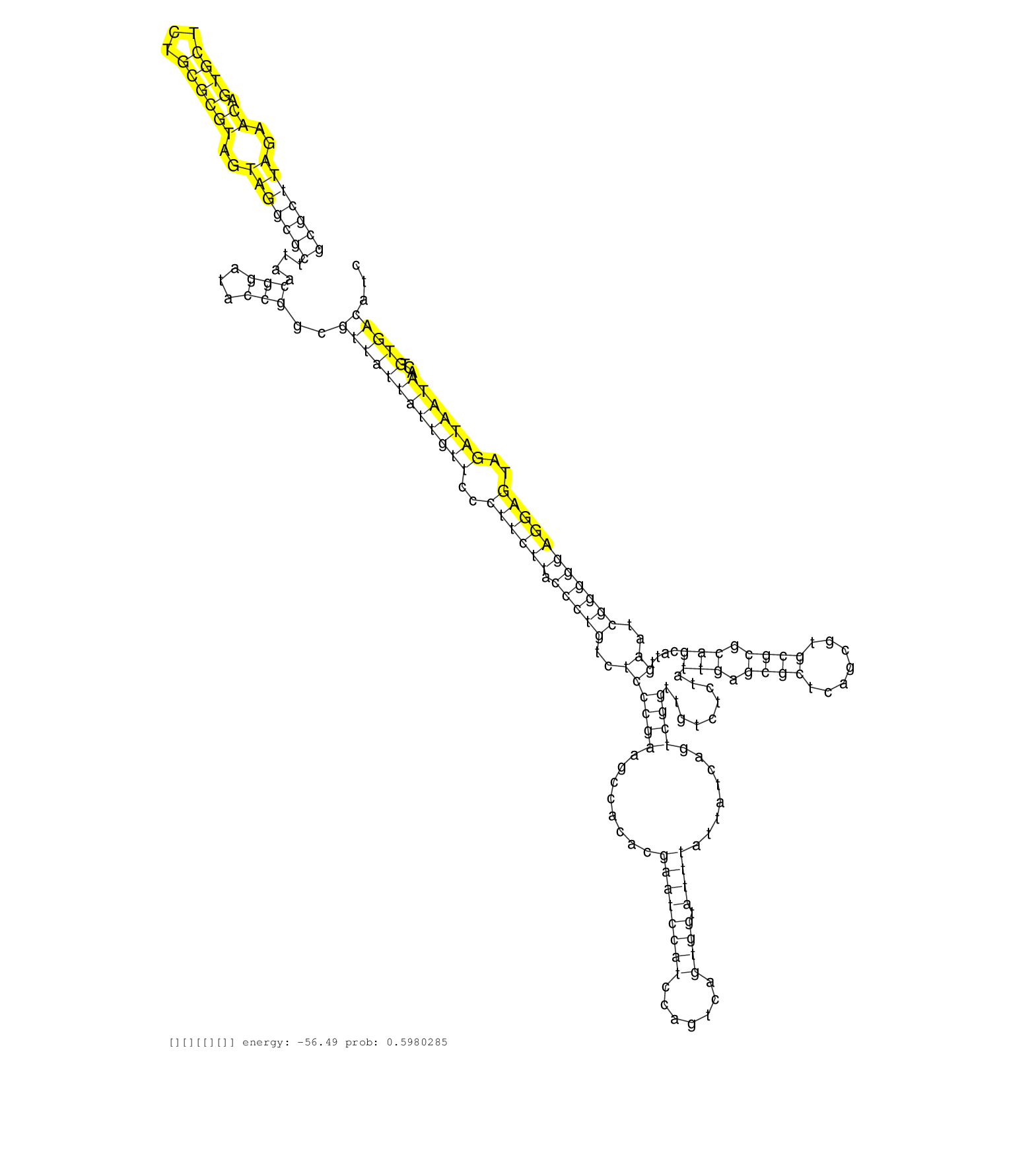
| TCCTAATGGCCAGGTTGAGTTTTGCGGGGGGCGTACTGACGCGACGGGACCACCTGGTGACCCTGTATCTCCCCCAGCGCTTAGAACAGTGCTCTGCGCGTAGTAGGCGCTTAACGGATACCGGCGTTATTATTGTTCCCTTCTTACCCTGTCTCCCGAAGCCACACGAATCCATCCAGTCAGTGGTATTTATTATCAGTCGGTTGTCTCTATTGAGCGCTCAGCGTGCGCGCAGCATTGAATCGGGGGAGGAGTAGATAATAACTGTGACATCTGTGCATCGCTTACCTCGTGCCGGGCACCGTTCTAAGCGCCGAGGTAGATAGGGGAAGATAATCAGGTTGGACACA ............................................................................(((((((..((.((((...))))))..)))))))....(((...)))..((((((((((((..((((((.(((((..((((((........((((((((.......)))).))))........)))).........(((.((((.......)))).)))....))..)))))))))))..)))))))...)))))............................................................................... ............................................................................77...................................................................................................................................................................................................274.......................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|
| .................................................................................TAGAACAGTGCTCTGCGCGTAGTGT.................................................................................................................................................................................................................................................... | 25 | 12.00 | 0.00 | 12.00 | - | |
| ................................................................................TTAGAACAGTGCTCTGCGCGTAGTGAGT.................................................................................................................................................................................................................................................. | 28 | 5.00 | 0.00 | 5.00 | - | |
| .................................................................................TAGAACAGTGCTCTGCGCGTAGTGAAA.................................................................................................................................................................................................................................................. | 27 | 2.00 | 0.00 | 2.00 | - | |
| ................................................................................TTAGAACAGTGCTCTGCGCGTAGTGT.................................................................................................................................................................................................................................................... | 26 | 2.00 | 0.00 | 2.00 | - | |
| .................................................................................TAGAACAGTGCTCTGCGCGTACTG..................................................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | |
| ..............................................................................GCTTAGAACAGTGCTCTGCGCGTAAA...................................................................................................................................................................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | |
| ...............................................................................CTTAGAACAGTGCTCTGCGCGTAGA...................................................................................................................................................................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | |
| ................................................................................TTAGAACAGTGCTCTGCGCGTAGTT..................................................................................................................................................................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | |
| .........................................................................................................................................................................................................................................................AGGAGTAGATAATAACTGTGA................................................................................ | 21 | 1 | 1.00 | 1.00 | - | 1.00 |
| .................................................................................TAGAACAGTGCTCTGCGCGTAGTGATC.................................................................................................................................................................................................................................................. | 27 | 1.00 | 0.00 | 1.00 | - | |
| .................................................................................TAGAACAGTGCTCTGCGCGTAGA...................................................................................................................................................................................................................................................... | 23 | 1.00 | 0.00 | 1.00 | - | |
| ................................................................................TTAGAACAGTGCTCTGCGCGTAGTCAGC.................................................................................................................................................................................................................................................. | 28 | 1.00 | 0.00 | 1.00 | - | |
| .................................................................................TAGAACAGTGCTCTGCGCGTAGTAGC................................................................................................................................................................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | |
| ................................................................................TTAGAACAGTGCTCTGCGCGTAGA...................................................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | |
| ................................................................................TTAGAACAGTGCTCTGCGCGTAGTTAA................................................................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | 1.00 | - | |
| .................................................................................TAGAACAGTGCTCTGCGCGTAGTGAGT.................................................................................................................................................................................................................................................. | 27 | 1.00 | 0.00 | 1.00 | - |
| TCCTAATGGCCAGGTTGAGTTTTGCGGGGGGCGTACTGACGCGACGGGACCACCTGGTGACCCTGTATCTCCCCCAGCGCTTAGAACAGTGCTCTGCGCGTAGTAGGCGCTTAACGGATACCGGCGTTATTATTGTTCCCTTCTTACCCTGTCTCCCGAAGCCACACGAATCCATCCAGTCAGTGGTATTTATTATCAGTCGGTTGTCTCTATTGAGCGCTCAGCGTGCGCGCAGCATTGAATCGGGGGAGGAGTAGATAATAACTGTGACATCTGTGCATCGCTTACCTCGTGCCGGGCACCGTTCTAAGCGCCGAGGTAGATAGGGGAAGATAATCAGGTTGGACACA ............................................................................(((((((..((.((((...))))))..)))))))....(((...)))..((((((((((((..((((((.(((((..((((((........((((((((.......)))).))))........)))).........(((.((((.......)))).)))....))..)))))))))))..)))))))...)))))............................................................................... ............................................................................77...................................................................................................................................................................................................274.......................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|