| (1) HEART | (1) KIDNEY | (1) OTHER |
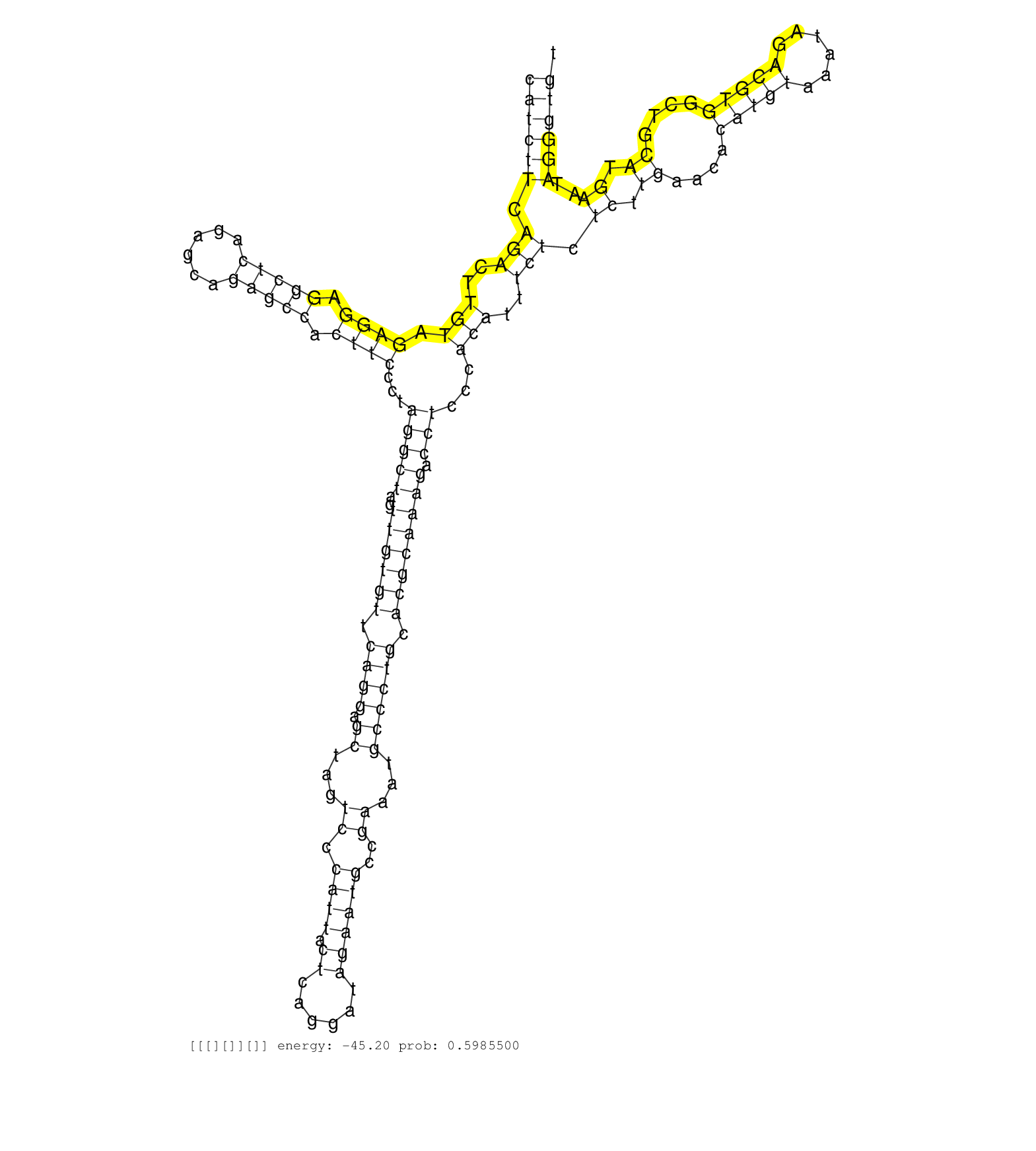
| CTGGAGTCAGAACCTCTCGCAGTTCTTTCCTGAGGCCATTACTTTCATTGGTAGGTGTTCGAGTTTTTGCTGCTTGTTTTACATTGAGGCCCTCCCAGTGCTCACACCCTACCAGGCATCTTCAGACTTGTAGAGGAGGCTCAGAGCAGAGCCACTTCCCTAGGCTAGTTGTGTTCAGGAGCTAGTCCCATTACTCAGGATAGAATGCCGAAATGCCCTGCACGCAAAGACCTCCCACATTTCTCTCTTGAACACATGTAAATAGACGTGGCTGCATGAATAGGGTGTCTGGGGCCTTTTGAGTTTGCCGTGACTGCGATGTCTGCACCTGTCTCCTTAAGACTGTACCT ....................................................................................................................((((((.(((..(((.((((.(((((......))))).))))...(((((..((((((.((((.((...((.((((.((......))))))..))...)))))).)))))))).)))...)))..))).((.((....(((((......)))))....)).))..))))))............................................................... ....................................................................................................................117........................................................................................................................................................................288............................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|
| ................................................................................................................CAGGCATCTTCAGACTTGTAGAGGAG.................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| .....................................................................................................................................................................................................................................................................................................................GTGACTGCGATGTCTGCACCTGTC................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .....................................................................................................................ATCTTCAGACTTGTAGAGGAGGCTCAGAG............................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .........................................................................................................................TCAGACTTGTAGAGGAG.................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .........................................................................................................................TCAGACTTGTAGAGGAGGCTCA............................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .......................................................................................................................................................................................................................................................................AGACGTGGCTGCATGAATAGG.................................................................. | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .....................................................................................................................................................................................................................................................TCTTGAACACATGTAAATAGACGT................................................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .....................................................................................................................ATCTTCAGACTTGTAGAGGA..................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .................................................................................................................AGGCATCTTCAGACTTGTAGAGG...................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .........................................................................................................................TCAGACTTGTAGAGGAGGCT................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .................................................................................................................AGGCATCTTCAGACTTGTAG......................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .................................................................................................................................................................AGGCTAGTTGTGTTCAGGAGC........................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .....................................ATTACTTTCATTGGTAGGTGTTCGAGTTTTTGCT....................................................................................................................................................................................................................................................................................... | 34 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..........................................................................................................................CAGACTTGTAGAGGA..................................................................................................................................................................................................................... | 15 | 2 | 0.50 | 0.50 | - | 0.50 | - |
| CTGGAGTCAGAACCTCTCGCAGTTCTTTCCTGAGGCCATTACTTTCATTGGTAGGTGTTCGAGTTTTTGCTGCTTGTTTTACATTGAGGCCCTCCCAGTGCTCACACCCTACCAGGCATCTTCAGACTTGTAGAGGAGGCTCAGAGCAGAGCCACTTCCCTAGGCTAGTTGTGTTCAGGAGCTAGTCCCATTACTCAGGATAGAATGCCGAAATGCCCTGCACGCAAAGACCTCCCACATTTCTCTCTTGAACACATGTAAATAGACGTGGCTGCATGAATAGGGTGTCTGGGGCCTTTTGAGTTTGCCGTGACTGCGATGTCTGCACCTGTCTCCTTAAGACTGTACCT ....................................................................................................................((((((.(((..(((.((((.(((((......))))).))))...(((((..((((((.((((.((...((.((((.((......))))))..))...)))))).)))))))).)))...)))..))).((.((....(((((......)))))....)).))..))))))............................................................... ....................................................................................................................117........................................................................................................................................................................288............................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|