| (1) HEART | (2) OTHER |
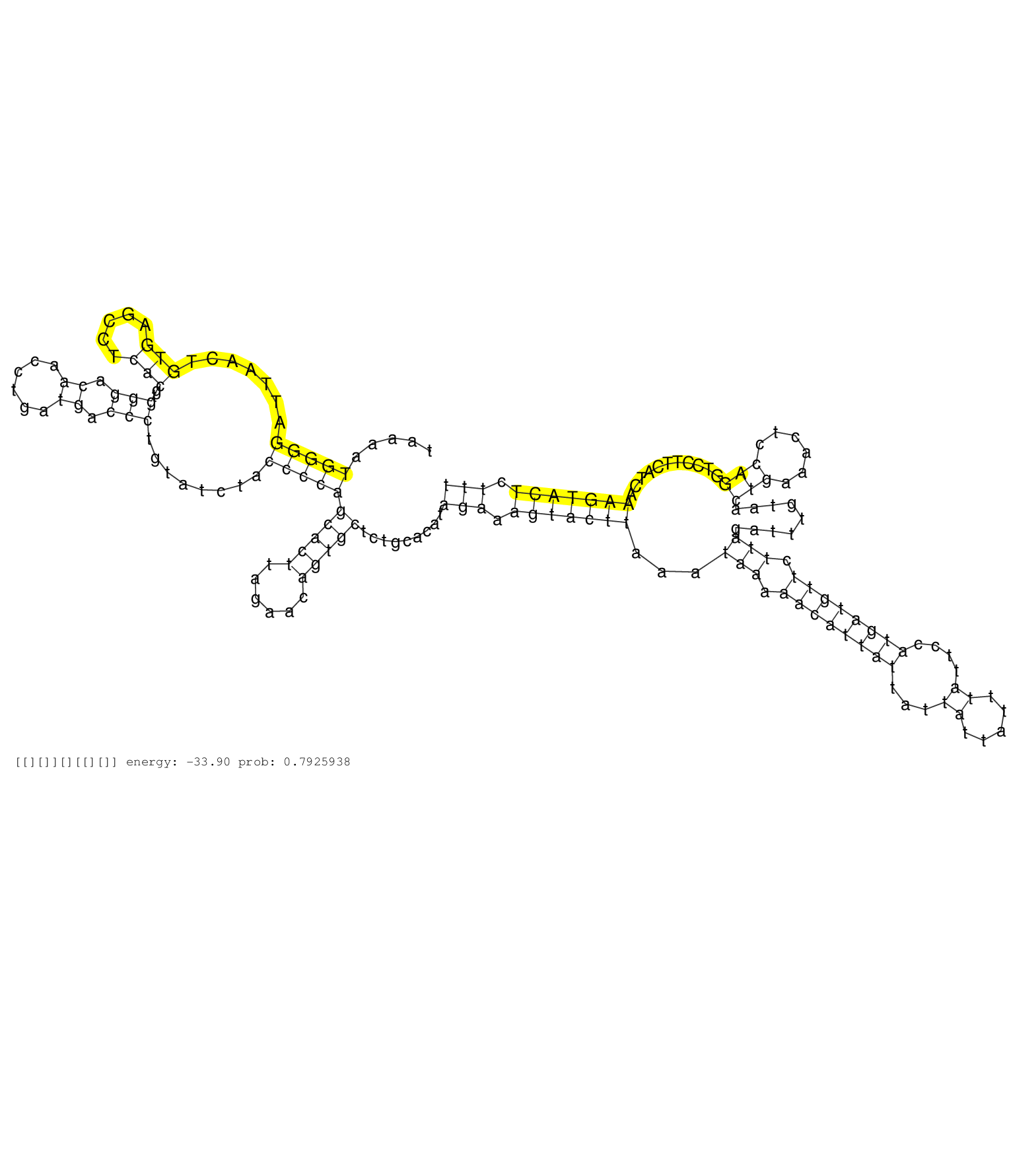
| CCCGGGCTTGGGAGTCAGGGGTCATGGGTTCGAATCCCAGATCTGCCACTTGTCAGCTGTGTGATTGTGGGCAAGTCACTTCACTTCTCTGTGCCTTAGTTACCTCATCTGTAAAATGGGGATTAACTGTGAGCCTCACGTGGGACAACCTGATGACCCTGTATCTACCCCAGCACTTAGAACAGTGCTCTGCACATAGAAAGTACTTAAATAAAAACATTATTATTATTATTTATTCCATGATGTTCTTAGATTTGTAACTGAAACTCCAGGTCCTTCATCAAAGTACTCTTTTCTTAGGGATTTAAACAAAATAGATGACTTGATGCAAGAGGTCACAGAACAACAAG ....................................................................................................................(((((.......(((.....)))..(((.((......)).)))........)))))(((((......))))).........(((.(((((((...(((.((((((((...((.....))....)))))))).))).........(((......)))...........))))))).)))........................................................ ...............................................................................................................112...................................................................................................................................................................................294...................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|
| ........................................................................................................TCATCTGTAAAATGGGGATTAACTGTGTAAA....................................................................................................................................................................................................................... | 31 | 1.00 | 0.00 | 1.00 | - | - | |
| ........................................................................................................TCATCTGTAAAATGGGGATTAACTGTGTTA........................................................................................................................................................................................................................ | 30 | 1.00 | 0.00 | 1.00 | - | - | |
| .............................................CCACTTGTCAGCTGTGTGATTGTGGGCATT................................................................................................................................................................................................................................................................................... | 30 | 1.00 | 0.00 | 1.00 | - | - | |
| ..............................................................................................................................................................................................................................................................................AGGTCCTTCATCAAAGTACT............................................................ | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ....................................................................................................................TGGGGATTAACTGTGAGGAA...................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | |
| ................................................................................................................................................................................TTAGAACAGTGCTCTGCACATAGATTGC.................................................................................................................................................. | 28 | 1.00 | 0.00 | 1.00 | - | - | |
| .................................................................................................................................................................................TAGAACAGTGCTCTGCACATAGAAGA................................................................................................................................................... | 26 | 1.00 | 0.00 | - | - | 1.00 | |
| .................................................................................................................................................................................TAGAACAGTGCTCTGCACATAGATTGC.................................................................................................................................................. | 27 | 1.00 | 0.00 | 1.00 | - | - | |
| ....................................................................................................................TGGGGATTAACTGTGAGGAAA..................................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | |
| .................................................................................................................................................................................TAGAACAGTGCTCTGCACATAGATTG................................................................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | - | |
| .............................................................................................................TGTAAAATGGGGATTAACTGTGAGCGTA..................................................................................................................................................................................................................... | 28 | 1.00 | 0.00 | 1.00 | - | - | |
| ........................................................................................................TCATCTGTAAAATGGGGATTAACTGTTATC........................................................................................................................................................................................................................ | 30 | 1.00 | 0.00 | 1.00 | - | - | |
| ..........................................................................................................................................................................................................................................................................................................................................AGAGGTCACAGAACAAC... | 17 | 2 | 0.50 | 0.50 | - | 0.50 | - |
| CCCGGGCTTGGGAGTCAGGGGTCATGGGTTCGAATCCCAGATCTGCCACTTGTCAGCTGTGTGATTGTGGGCAAGTCACTTCACTTCTCTGTGCCTTAGTTACCTCATCTGTAAAATGGGGATTAACTGTGAGCCTCACGTGGGACAACCTGATGACCCTGTATCTACCCCAGCACTTAGAACAGTGCTCTGCACATAGAAAGTACTTAAATAAAAACATTATTATTATTATTTATTCCATGATGTTCTTAGATTTGTAACTGAAACTCCAGGTCCTTCATCAAAGTACTCTTTTCTTAGGGATTTAAACAAAATAGATGACTTGATGCAAGAGGTCACAGAACAACAAG ....................................................................................................................(((((.......(((.....)))..(((.((......)).)))........)))))(((((......))))).........(((.(((((((...(((.((((((((...((.....))....)))))))).))).........(((......)))...........))))))).)))........................................................ ...............................................................................................................112...................................................................................................................................................................................294...................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|