| (2) OTHER |
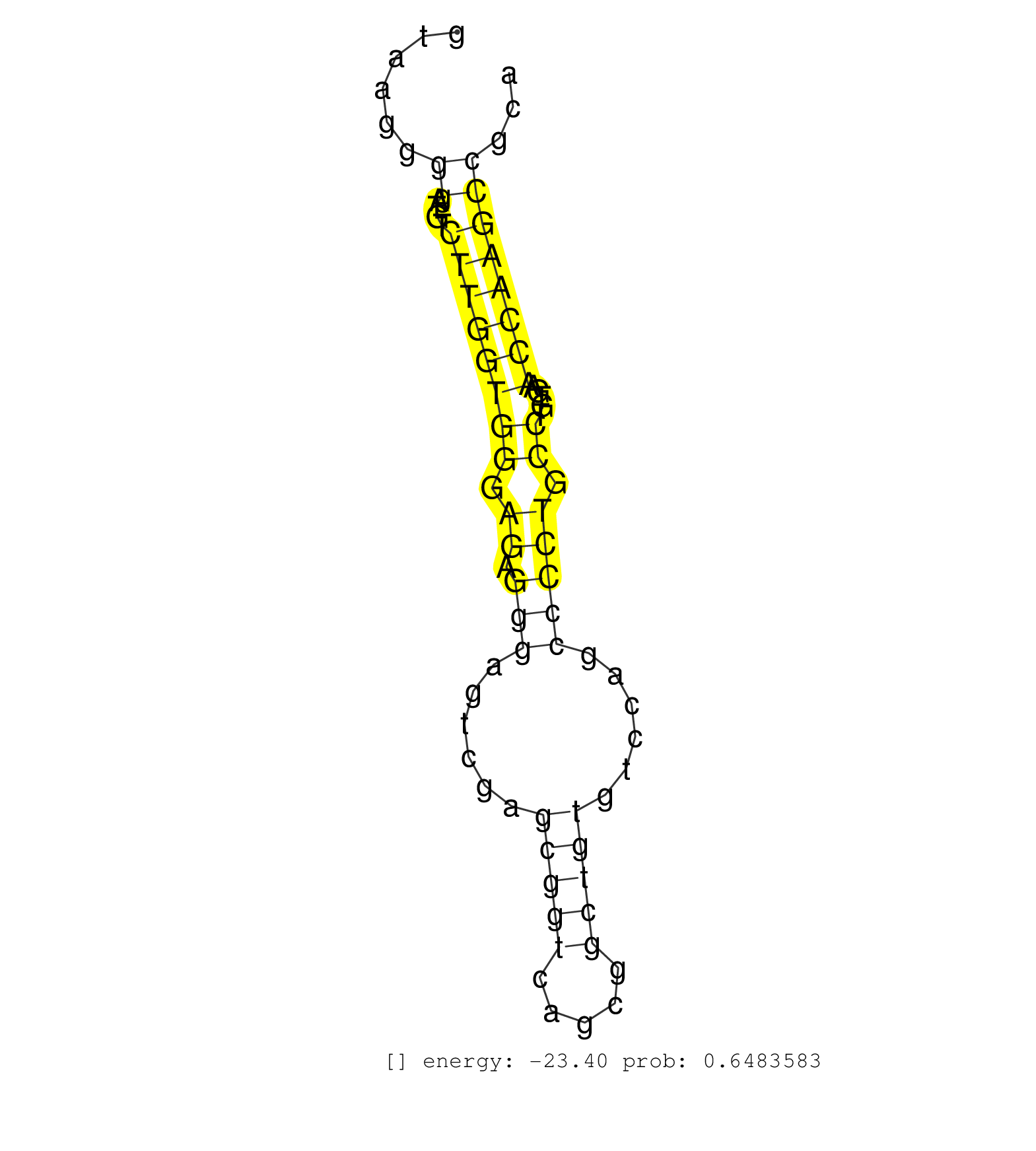
| TGGGCAGCCTGGCTTGGCCACTCACTCATCCGTTAACAGGGTCCTGGCAGGTAAGGGGATGTCTTGGTGGGAGAGGGAGTCGAGCGGTCAGCGGCTGTGTCCAGCCCCTGCCTGTGAACCAAGCCGCAGAACGGAGAGAGCCTGAGAACGTCAGGCTTGTAGGGAAGAGTAAAACCTGCTCTTTGACTCTGATTTGAAAGCCCTGCTCCCTCCCCACCTAACCCTCCCCGCTTTTGAAACCTCAAATTGACTGTTTCATAATGTACCGATTAGATTCCTGGCCAGGCTTCTAATCAAATAAGGTATCTTGTTCACGTAATGGTTGGTGAAATGCTCACGAGGCATATCCT ........................................................((....((((((((.((.(((......(((((.....)))))......))))).)).....))))))))................................................................................................................................................................................................................................. ..................................................51...........................................................................128............................................................................................................................................................................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|
| ..........................................................................................................CCTGCCTGTGAACCAAGC.................................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | 1.00 | - |
| ..........................................................ATGTCTTGGTGGGAGAG................................................................................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | 1.00 | - |
| .....................................................................................................................................................GTCAGGCTTGTAGGGAAG....................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - |
| .................................................................................................................GTGAACCAAGCCGCAGAA........................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - |
| ..................................................................................................................................................AACGTCAGGCTTGTAGGGAAGAGTAA.................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - |
| ..................................................................................................................................ACGGAGAGAGCCTGAAA........................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | 1.00 | - | |
| ...............................................................................................................................................GAGAACGTCAGGCTTGTAG............................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | 1.00 | - |
| .......................................................................................................................................................CAGGCTTGTAGGGAAGAG..................................................................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | 0.50 | - |
| .......................................................................................................................................................CAGGCTTGTAGGGAA........................................................................................................................................................................................ | 15 | 8 | 0.12 | 0.12 | 0.12 | - |
| TGGGCAGCCTGGCTTGGCCACTCACTCATCCGTTAACAGGGTCCTGGCAGGTAAGGGGATGTCTTGGTGGGAGAGGGAGTCGAGCGGTCAGCGGCTGTGTCCAGCCCCTGCCTGTGAACCAAGCCGCAGAACGGAGAGAGCCTGAGAACGTCAGGCTTGTAGGGAAGAGTAAAACCTGCTCTTTGACTCTGATTTGAAAGCCCTGCTCCCTCCCCACCTAACCCTCCCCGCTTTTGAAACCTCAAATTGACTGTTTCATAATGTACCGATTAGATTCCTGGCCAGGCTTCTAATCAAATAAGGTATCTTGTTCACGTAATGGTTGGTGAAATGCTCACGAGGCATATCCT ........................................................((....((((((((.((.(((......(((((.....)))))......))))).)).....))))))))................................................................................................................................................................................................................................. ..................................................51...........................................................................128............................................................................................................................................................................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................................................................................................TACGTGAACAAGATACCTTATTTGATTAGA................................ | 30 | 1 | 1.00 | 1.00 | - | 1.00 |