| (1) HEART | (1) KIDNEY | (1) OTHER |
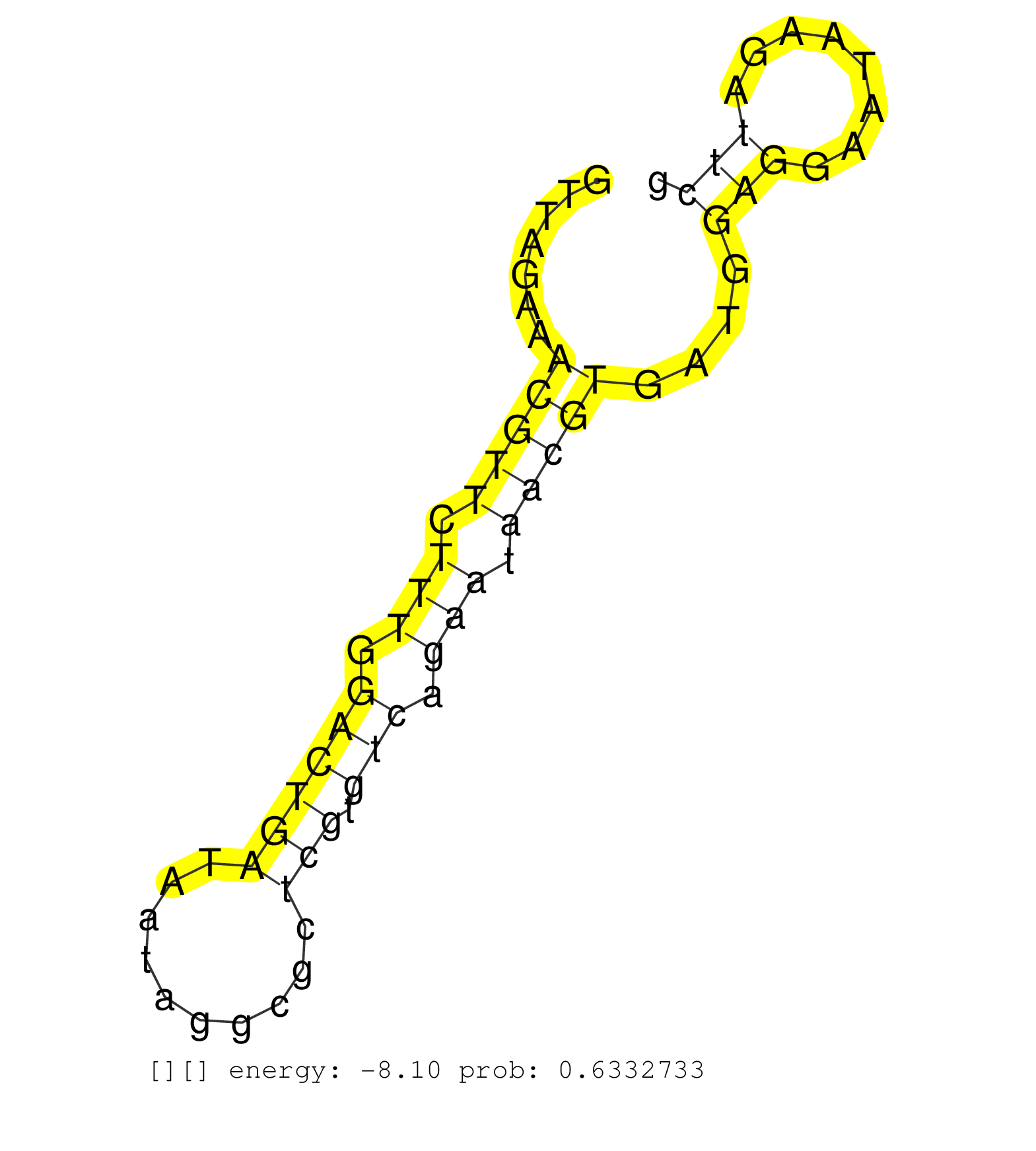
| CGCTCAAGAGGACGTCCGTCCCGCCGAGACCCAGTCCGGGATGTGGGAAGGTTAGAAACGTTCTTTGGACTGATAATAGGCGCTCGTGTCAGAATAACGTGATGGAGGAATAAGATTCGCCAAAATTTTGCCCTGCGGTCAAGAAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAAAAAAAAAAAAACCAAATCCTCAAATCAAGCCGCATGGACTGGTTTGTGGCTTCCTTGAGGATTTCCCAAGGTCACTCCCAGTCTCGGGCATTTTTTTCAAGAGGAAAATGGGGATCTTGCCTTATGCAGAAAATAAAGGCCGAGGAGTCGATGAGGAGTGTCCGCTTA .........................................................(((((.(((.((((((..........))).))).))).)))))....(((........)))........................................................................................................................................................................................................................................ ..................................................51..................................................................119..................................................................................................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| .................................GTCCGGGATGTGGGAAG............................................................................................................................................................................................................................................................................................................ | 17 | 1 | 6.00 | 6.00 | 2.00 | 4.00 | - |
| ..................................................GTTAGAAACGTTCTTTGGACTGATA................................................................................................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 2.00 | - | - |
| ..CTCAAGAGGACGTCCGT........................................................................................................................................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ...............................................................................................................................................................................................................................................................................................................................GGCCGAGGAGTCGATGAGGAGTGTCCGC... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...............................................................TTTGGACTGATAATAGGCGCT.......................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ..................................TCCGGGATGTGGGAAGGT.......................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .................................................GGTTAGAAACGTTCTTTGGACT....................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ...............................................................TTTGGACTGATAATAGGCGC........................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ......................................................................................................................................................................................................................................................................................................................GAAAATAAAGGCCGAGGAGTCGATG............... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ..................................................................................................GTGATGGAGGAATAAGA........................................................................................................................................................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | 0.50 |
| ..........GACGTCCGTCCCGCC..................................................................................................................................................................................................................................................................................................................................... | 15 | 2 | 0.50 | 0.50 | - | - | 0.50 |
| ...................................CCGGGATGTGGGAAG............................................................................................................................................................................................................................................................................................................ | 15 | 8 | 0.25 | 0.25 | - | 0.25 | - |
| ................................................................................................................................................................................................................................................................................................................TATGCAGAAAATAAA............................... | 15 | 14 | 0.07 | 0.07 | - | 0.07 | - |
| CGCTCAAGAGGACGTCCGTCCCGCCGAGACCCAGTCCGGGATGTGGGAAGGTTAGAAACGTTCTTTGGACTGATAATAGGCGCTCGTGTCAGAATAACGTGATGGAGGAATAAGATTCGCCAAAATTTTGCCCTGCGGTCAAGAAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAAAAAAAAAAAAACCAAATCCTCAAATCAAGCCGCATGGACTGGTTTGTGGCTTCCTTGAGGATTTCCCAAGGTCACTCCCAGTCTCGGGCATTTTTTTCAAGAGGAAAATGGGGATCTTGCCTTATGCAGAAAATAAAGGCCGAGGAGTCGATGAGGAGTGTCCGCTTA .........................................................(((((.(((.((((((..........))).))).))).)))))....(((........)))........................................................................................................................................................................................................................................ ..................................................51..................................................................119..................................................................................................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|