| (1) HEART | (1) KIDNEY | (2) OTHER |
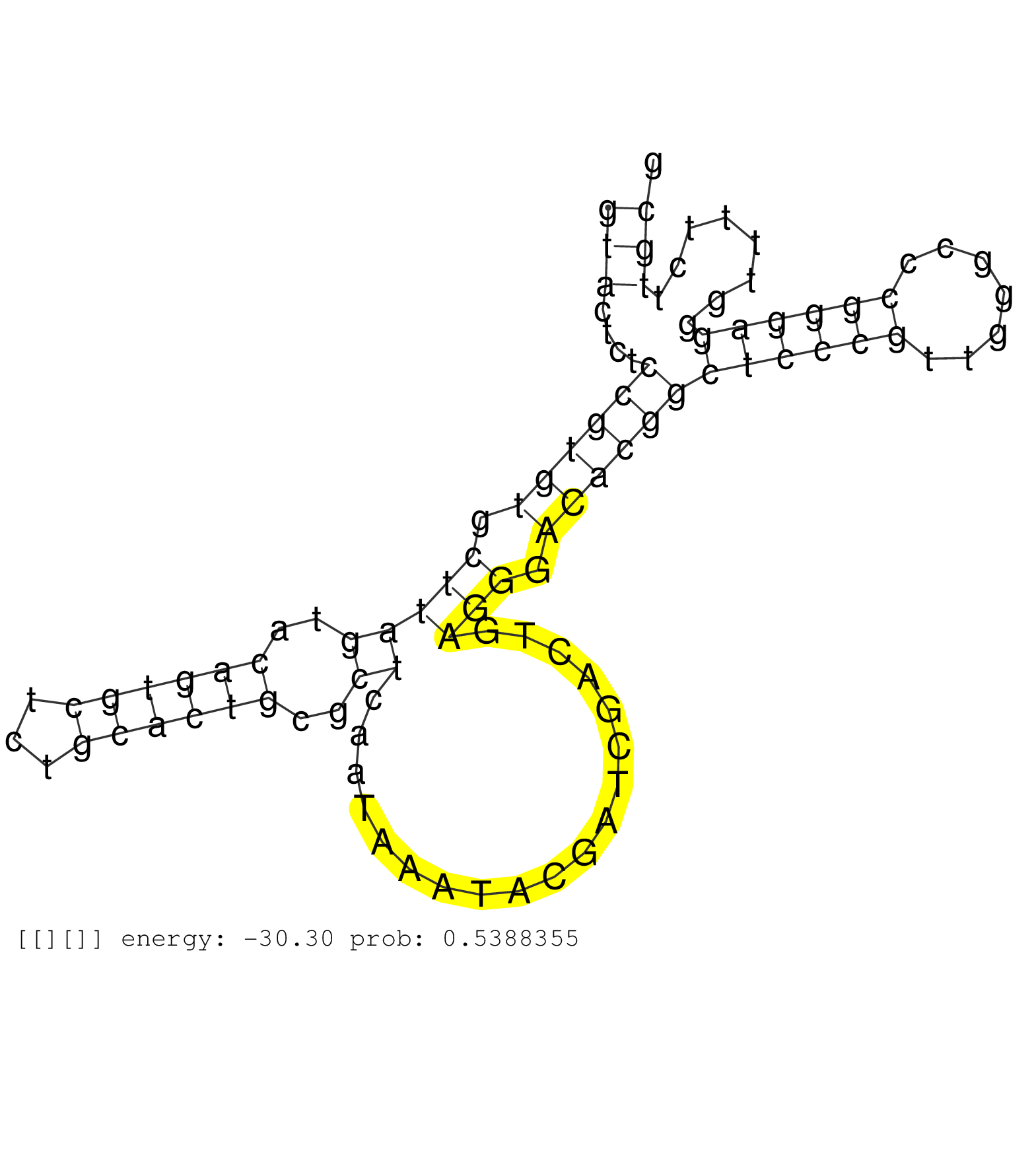
| TGGGGCCCGCGGGGAACCGAAGCCCGTTGTGCCCGGGGAGGGGTGGCACGGGATGGCTCTGCCGGGCCGGCGCCCCCGACCCGTGATGGGAAGGCCGGGGCCGGTAGAGACGAGGCTGCGGGAACAGCCGGGCTCCCGGGGCTGCCCCTGGCCCGCTGCAGGAAATCCCGTCGTTGCGGGCAGGGAATGTGTGTTCTATTGTACTCTCCGTGTGCTTAGTACAGTGCTCTGCACTGCGCTCAATAAATACGATCGACTGAGGGACACGGCTCCCGTTGGGCCCGGGAGGGTTTTCTTGCGGGCACGATTCAGTGGGGCCAGACGGAAGGGGGATGGATCCCCCTGATCCC ........................................................................................................................................................................................................(((....((((((.(((((..((((((...))))))..))...................))).))))))((((((.......))))))........)))................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................................AATAAATACGATCGACTGAGGGACACGGCTCC............................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| .......................................................................................................................................................................................................................................................TACGATCGACTGAGGGACACGGC................................................................................ | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................................................................GACTGAGGGACACGGCTCCCGTTGGGCC.................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ....................................................................................................................................................................................................................................................................................................................TCAGTGGGGCCAGACAG......................... | 17 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ...................................................................................................................................................................................................................................................TAAATACGATCGACTGAGTGAG..................................................................................... | 22 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ......................................................................................................................................................................................................................................................ATACGATCGACTGAGGGACACGG................................................................................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .......................................................................................................................................................................................................................................................TACGATCGACTGAGGGACACTGCT............................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ...............................................................................................................................................................................................................................................TCAATAAATACGATCGACTGAGGGACA.................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ......................................................................................................................................................................................................................................................ATACGATCGACTGAGGGACACGGCTCCCCTTG........................................................................ | 32 | 1.00 | 0.00 | - | - | - | 1.00 | |
| ......................................................................................................................................................................................................................................................ATACGATCGACTGAGGGACACGGC................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ..........................................................................................................................................................................................................................................................GATCGACTGAGGGACA.................................................................................... | 16 | 2 | 0.50 | 0.50 | 0.50 | - | - | - |
| ...................................................................................................................................................................................................................................................TAAATACGATCGACTGAGG........................................................................................ | 19 | 10 | 0.10 | 0.10 | - | 0.10 | - | - |
| TGGGGCCCGCGGGGAACCGAAGCCCGTTGTGCCCGGGGAGGGGTGGCACGGGATGGCTCTGCCGGGCCGGCGCCCCCGACCCGTGATGGGAAGGCCGGGGCCGGTAGAGACGAGGCTGCGGGAACAGCCGGGCTCCCGGGGCTGCCCCTGGCCCGCTGCAGGAAATCCCGTCGTTGCGGGCAGGGAATGTGTGTTCTATTGTACTCTCCGTGTGCTTAGTACAGTGCTCTGCACTGCGCTCAATAAATACGATCGACTGAGGGACACGGCTCCCGTTGGGCCCGGGAGGGTTTTCTTGCGGGCACGATTCAGTGGGGCCAGACGGAAGGGGGATGGATCCCCCTGATCCC ........................................................................................................................................................................................................(((....((((((.(((((..((((((...))))))..))...................))).))))))((((((.......))))))........)))................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|