| (1) HEART | (1) KIDNEY |
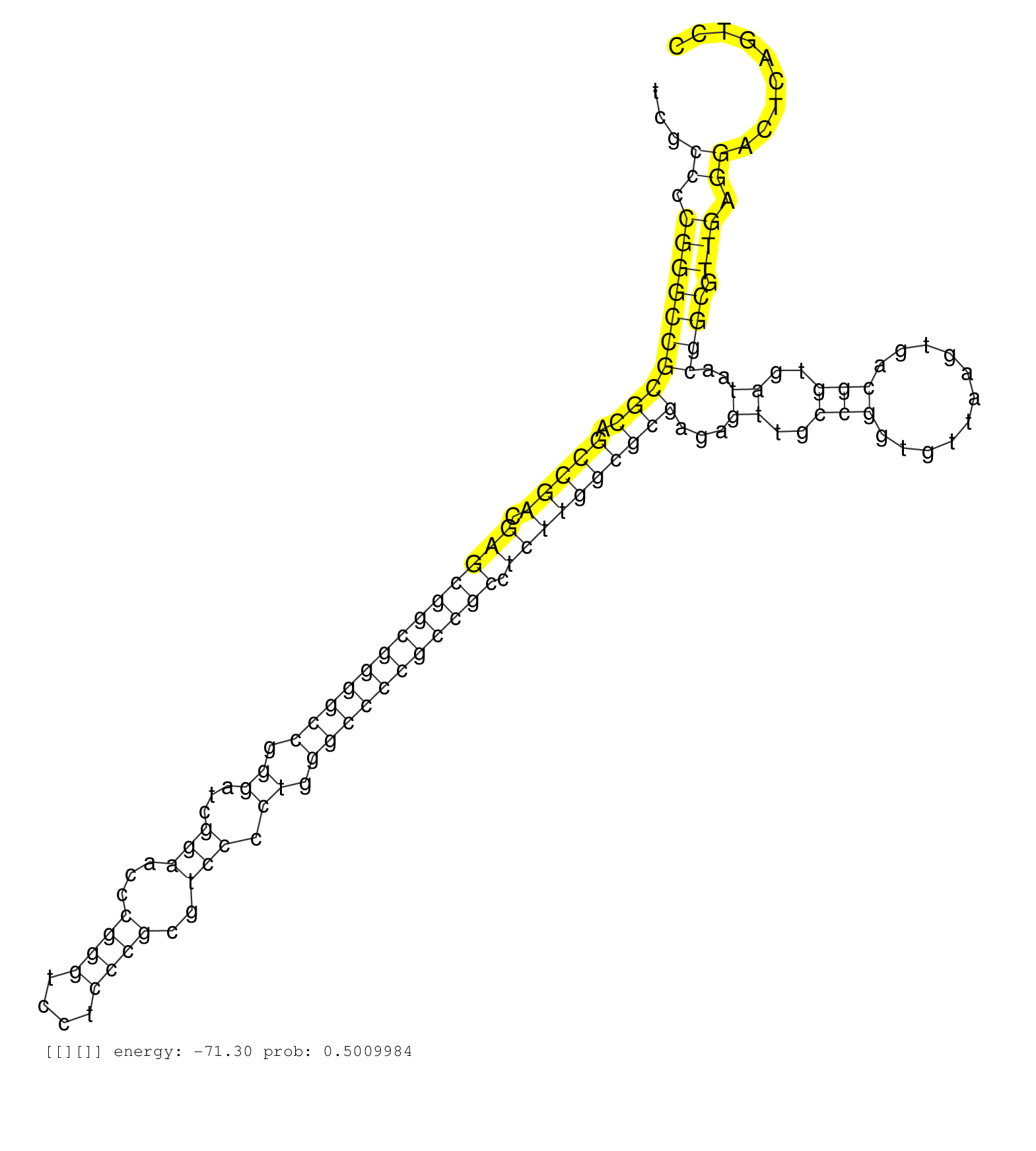
| GCGTTAATCCCCGTTTTACAGGGGAGGTGACCGAGGCTGAGTGAAGGCACTCGCCCCGGGCCGCGCAGCCGACGAGCGGCGGGGCCGGGATCGGAACCCGGGTCCTCCCGCGTCCCCTGGGCCCCGCCGCCTCTTGGCGCGAGAGTTGCCGGTGTTAAGTGACGGTGATAACGGCGTTGAGGACTCAGTCCGAGTTAAGCGCCGAATCAGCAGACGGTGATTCGGTGATACGCGGTGCCCGCGCCCCTCGCCCGCGGCCTCGACCCCGAGGCGGCCGGCGGACGTCCCCGTCTCCCTCAGGCACCTCCTGATCACCCAGCGGATCCTGCAGAACGTTCCGCCCGTCGTCT .....................................................((.((((((((((.(((((.(((((((((((((.((...(((...((((....))))..))).)).))))))))))).))))))))))...((..(((...........)))..))..)))).))).))........................................................................................................................................................................ ..................................................51..........................................................................................................................................191............................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................................................................................................................GCGGATCCTGCAGAA................. | 15 | 1 | 2.00 | 2.00 | 2.00 | - |
| ........................................................CGGGCCGCGCAGCCGACGAG.................................................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | 1.00 | - |
| ...............................................................................................................................................................................GTTGAGGACTCAGTCCGAGTTAAGC...................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - |
| .............................................................................................................................................................................................................................................................................................................................AGCGGATCCTGCAGAACGT.............. | 19 | 1 | 1.00 | 1.00 | 1.00 | - |
| ...........CGTTTTACAGGGGAGGTGACCGAGGCTGAG..................................................................................................................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - |
| ..............................................................................................................................................................................................................................................................................................................................GCGGATCCTGCAGAACG............... | 17 | 1 | 1.00 | 1.00 | 1.00 | - |
| ..............................................................................................................................................................................................................................................................................................................................GCGGATCCTGCAGAACGTCCC........... | 21 | 1.00 | 0.00 | 1.00 | - | |
| ..............................................................................................................................................................................................................................................................................................................................GCGGATCCTGCAGAACGTCC............ | 20 | 1.00 | 0.00 | 1.00 | - | |
| .............................................................................................................................................................................GCGTTGAGGACTCAGTCCGA............................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | 1.00 | - |
| .............................................................................................................................................................................................................................................................................................................................AGCGGATCCTGCAGAAC................ | 17 | 1 | 1.00 | 1.00 | - | 1.00 |
| ..............................................................................................................................................................GTGACGGTGATAACGGC............................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | 1.00 | - |
| ...................................................................................................................................................................................................TAAGCGCCGAATCAGCAGACG...................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - |
| ...........................................................................................................................................................................................................................................................................................................................CCAGCGGATCCTGCAGAACG............... | 20 | 1 | 1.00 | 1.00 | - | 1.00 |
| ..............................................................................................................................................................................................................................................................................................................................GCGGATCCTGCAGAAC................ | 16 | 1 | 1.00 | 1.00 | 1.00 | - |
| .............................................................................................................................................................................................................................................................................................................CACCTCCTGATCACCC................................. | 16 | 3 | 0.33 | 0.33 | 0.33 | - |
| GCGTTAATCCCCGTTTTACAGGGGAGGTGACCGAGGCTGAGTGAAGGCACTCGCCCCGGGCCGCGCAGCCGACGAGCGGCGGGGCCGGGATCGGAACCCGGGTCCTCCCGCGTCCCCTGGGCCCCGCCGCCTCTTGGCGCGAGAGTTGCCGGTGTTAAGTGACGGTGATAACGGCGTTGAGGACTCAGTCCGAGTTAAGCGCCGAATCAGCAGACGGTGATTCGGTGATACGCGGTGCCCGCGCCCCTCGCCCGCGGCCTCGACCCCGAGGCGGCCGGCGGACGTCCCCGTCTCCCTCAGGCACCTCCTGATCACCCAGCGGATCCTGCAGAACGTTCCGCCCGTCGTCT .....................................................((.((((((((((.(((((.(((((((((((((.((...(((...((((....))))..))).)).))))))))))).))))))))))...((..(((...........)))..))..)))).))).))........................................................................................................................................................................ ..................................................51..........................................................................................................................................191............................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|