| (1) HEART | (1) KIDNEY | (1) OTHER |
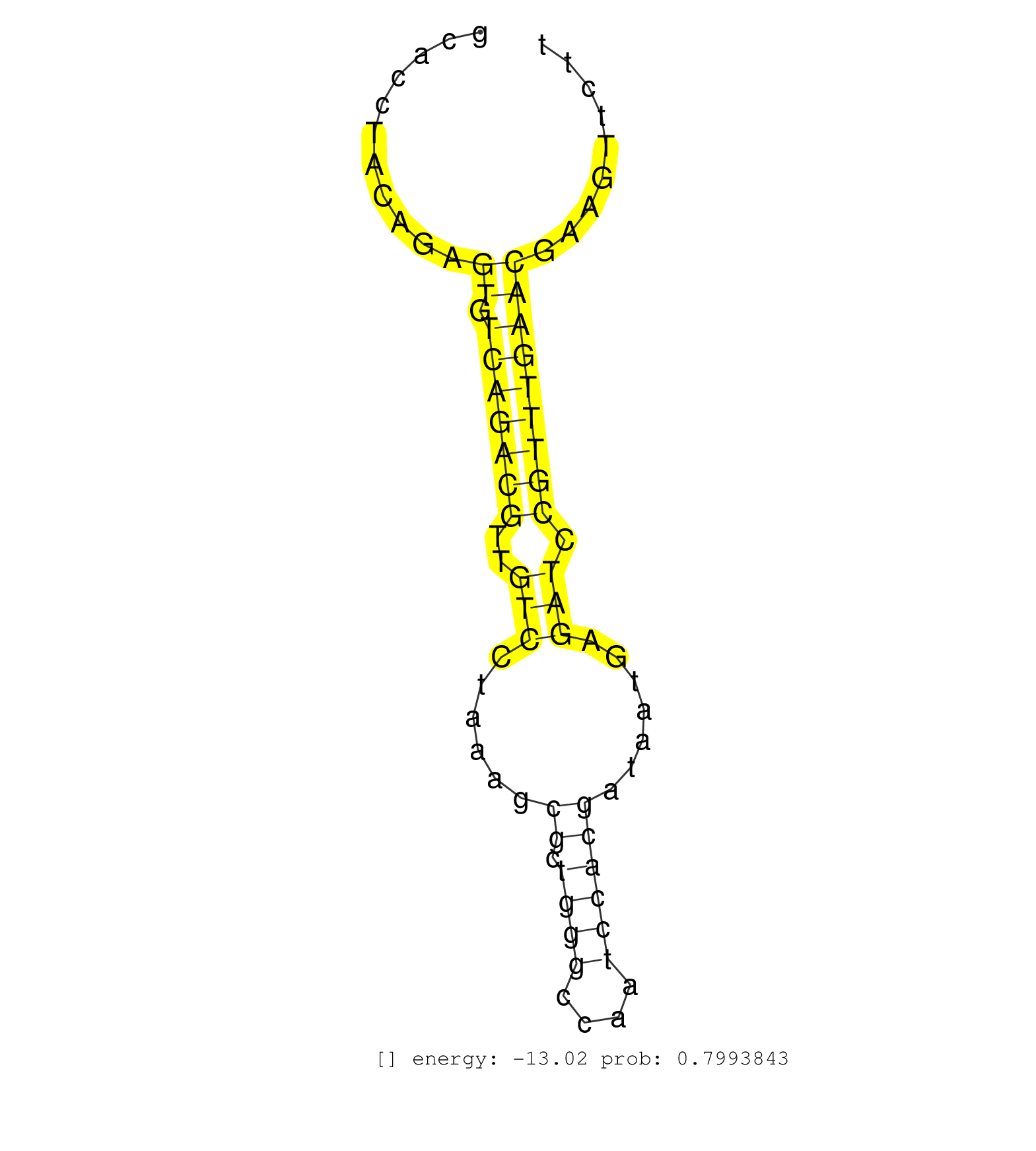
| GGAGGGCGGAGACCTCCAAGTTAACAGAACCCCATTTAGTTAGTGCGGGTATTTACGGAGCACCTACAGAGTGTCAGACGTTGTCCTAAAGCGCTGGGCCAATCCACGATAATGAGATCCGTTTGAACGAAGTTCTTCTTGGCCTTGCTGCCCCATCCACACATCAGAGAACGACGCACCCGTAGCCGTCAAGCGTGACCCTTTCCCCTCCGCCCGGGTGGCCAGCTTCGGCTCCTTTCGTGTGGAGAGAATGGTTATTACCAGGGAAGGTTTGTTTCTTTTGTTTTACTTGCTATCTAGATGCCGCACAGACTCTCGCACAGGAACAGAGCATGGATATTCCCAGCCAA ......................................................................((.(((((((..(((......((.((((....)))))).......))).))))))))).............................................................................................................................................................................................................................. ...........................................................60...........................................................................137................................................................................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|
| ................................................................TACAGAGTGTCAGACGTTGTCC........................................................................................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .....................................................................AGTGTCAGACGTTGTCCTAAAGCGCTGGGC........................................................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .................................................................................................................................................................................................................................................................................................................GCACAGACTCTCGCACAGGAACAGAGCATGG.............. | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ..........................................................................................................................................................................................................................................................................................................AGATGCCGCACAGACTCTCGCACAGG.......................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..................................................................................................................................................GCTGCCCCATCCACACATCAGAGAACGAC............................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ..................................................................................................................................................................................................................................................................................................................CACAGACTCTCGCACAG........................... | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .................................................................................................................GAGATCCGTTTGAACGAAGT......................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..................................................................................................................................................GCTGCCCCATCCACACATCAGAG..................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ...........................................................................................................................................................................................................................................................................................................GATGCCGCACAGACTCTCGCAC............................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..................................................................................................................................................................................................................................................................................................................CACAGACTCTCGCACAGGAACAG..................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| GGAGGGCGGAGACCTCCAAGTTAACAGAACCCCATTTAGTTAGTGCGGGTATTTACGGAGCACCTACAGAGTGTCAGACGTTGTCCTAAAGCGCTGGGCCAATCCACGATAATGAGATCCGTTTGAACGAAGTTCTTCTTGGCCTTGCTGCCCCATCCACACATCAGAGAACGACGCACCCGTAGCCGTCAAGCGTGACCCTTTCCCCTCCGCCCGGGTGGCCAGCTTCGGCTCCTTTCGTGTGGAGAGAATGGTTATTACCAGGGAAGGTTTGTTTCTTTTGTTTTACTTGCTATCTAGATGCCGCACAGACTCTCGCACAGGAACAGAGCATGGATATTCCCAGCCAA ......................................................................((.(((((((..(((......((.((((....)))))).......))).))))))))).............................................................................................................................................................................................................................. ...........................................................60...........................................................................137................................................................................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|