| (1) HEART | (1) KIDNEY | (1) OTHER |
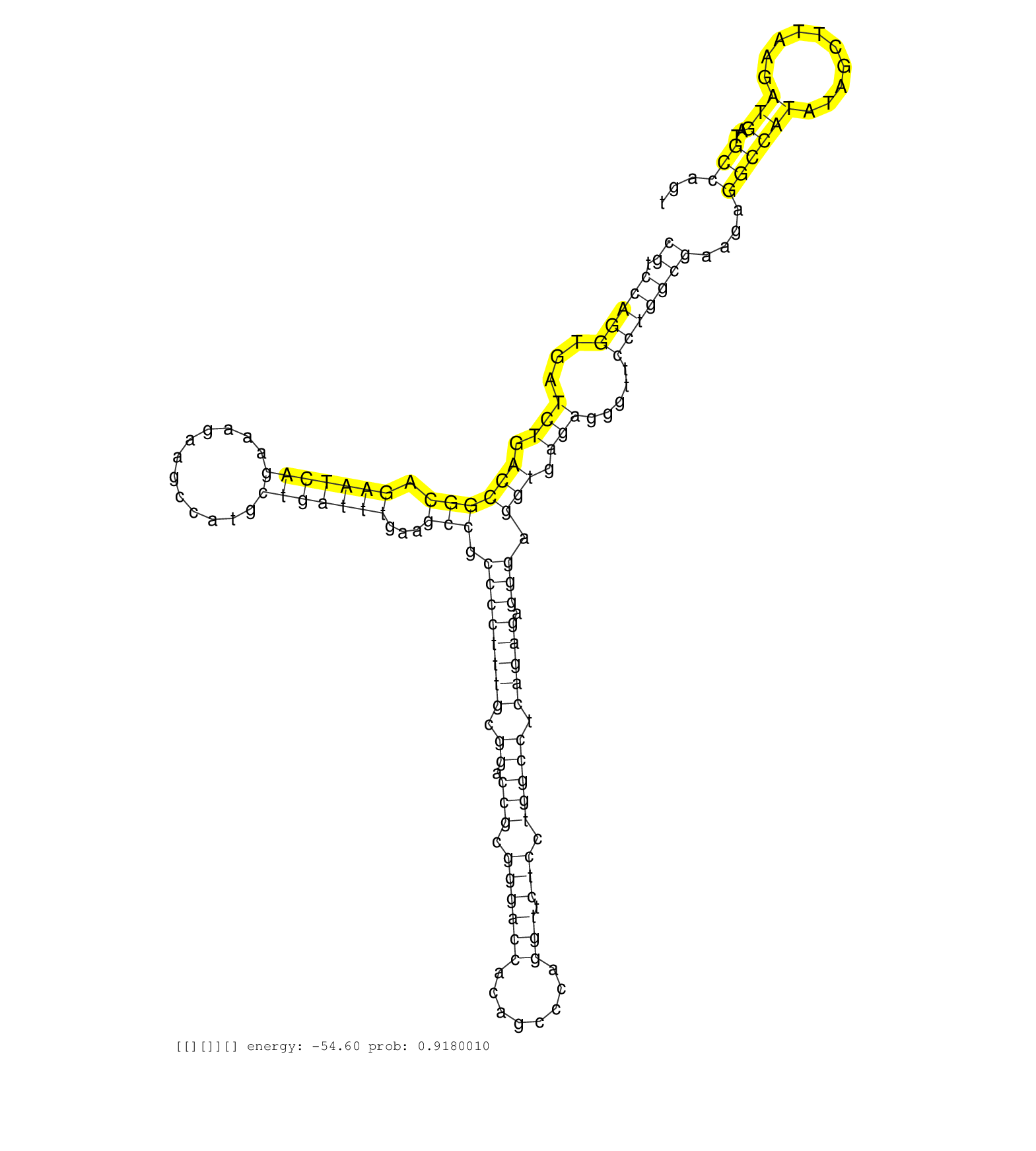
| CACTTCCCCTTTTCCCTCAGGATGTTGGCAGTCCTTGACCCAAAACCCGACTACCAGCACTACGTGGACACCTCCTGATGGAGCCGGGCTCTCCGGGAGCCGGTCGGAAGAGAGACAGTGAGATCCCGTCCAGGTGATCTGACCGGCAGAATCAGAAAGAAGCCATGCTGATTTGAAGCCGCCCCTTTGCGGACCGCGGGACCACAGCCCAGGTTCTCCTGGCCTCAGAGAGGGAGGTGAGAGGGTTCCTGGCGAAGAGGCCATATAGCTTAAGATGATGCCAGTGATTTTCTCAGGCAGCTGCACGAGGAGAGGGGCCAGCTATTTAGGCTTCTCTGAGCACCAGCAGG ..............................................................................................................................((.(((((...(((.((((((.(((((((............)))))))...))).((((((((.((.(((.((((((........))).))).))))).))))).))).))).))).....)))))))....((((((..........)))..))).................................................................... ..............................................................................................................................127...........................................................................................................................................................285............................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................................................................................................................................TATTTAGGCTTCTCTGAGCACCAGC... | 25 | 1 | 2.00 | 2.00 | - | 2.00 | - |
| ..................................................................................................................................................................................................................................................................GGCCATATAGCTTAAGATGATGC..................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...................................................................................................................................AGGTGATCTGACCGGCAGAATCA.................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .....................................................................................................GGTCGGAAGAGAGACAGTGA..................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ............................................................................................................................CCCGTCCAGGTGATCTGACCGGC........................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ...................................................................................................................................AGGTGATCTGACCGGCA.......................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..............................................................................................................................................CCGGCAGAATCAGAAAGAAGCCATGC...................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .....................................................................................................................................GTGATCTGACCGGCAGAATCA.................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ...................................................TACCAGCACTACGTGGACACCTCCTG................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| .........................................................................................................................GATCCCGTCCAGGTGATCTGACCGCCA.......................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | 1.00 | - | - | |
| ...................................................................................................CCGGTCGGAAGAGAGACAGTGAGATCC................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ....................................................................................................................................................................................................................................................................................GATGCCAGTGATTTTCTCA....................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .........................................................................................................................................................................GATTTGAAGCCGCCCC..................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .............................................................................................CGGGAGCCGGTCGGAAGAGA............................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| CACTTCCCCTTTTCCCTCAGGATGTTGGCAGTCCTTGACCCAAAACCCGACTACCAGCACTACGTGGACACCTCCTGATGGAGCCGGGCTCTCCGGGAGCCGGTCGGAAGAGAGACAGTGAGATCCCGTCCAGGTGATCTGACCGGCAGAATCAGAAAGAAGCCATGCTGATTTGAAGCCGCCCCTTTGCGGACCGCGGGACCACAGCCCAGGTTCTCCTGGCCTCAGAGAGGGAGGTGAGAGGGTTCCTGGCGAAGAGGCCATATAGCTTAAGATGATGCCAGTGATTTTCTCAGGCAGCTGCACGAGGAGAGGGGCCAGCTATTTAGGCTTCTCTGAGCACCAGCAGG ..............................................................................................................................((.(((((...(((.((((((.(((((((............)))))))...))).((((((((.((.(((.((((((........))).))).))))).))))).))).))).))).....)))))))....((((((..........)))..))).................................................................... ..............................................................................................................................127...........................................................................................................................................................285............................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|