| (1) HEART | (1) KIDNEY | (1) OTHER |
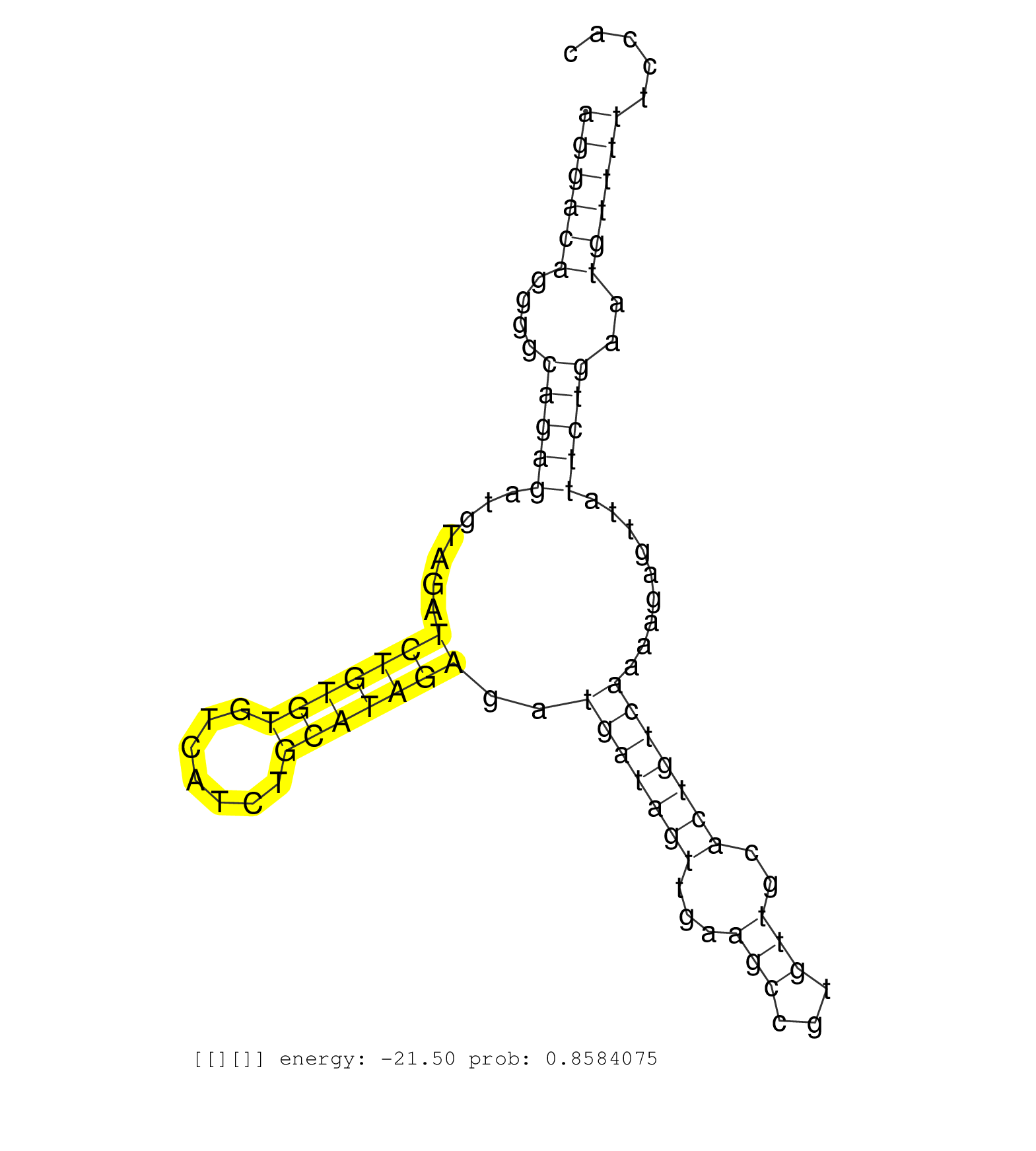
| ATGACACCGAGGTTGCGGGCCTGAGAGACGGGAAGGATGGTCGTACCATCCACGGTGATAGGGAAGTCAAGGAGAAGACAGGTCTTGGGAGGGAAGATGAGGAGCCCAGTTTTGGTCATGTTGACTTCTGGTGGCGGGCAGACATCCAGGTAGAGACATCTTGGAGGCAGGAGGAGATACGAGCCTGAAGGGAGGTGGAGAGGACAGGGGCAGAGATGTAGATCTGTGTGTCATCTGCATAGAGATGATAGTTGAAGCCGTGTTGCACTGTCAAAAGAGTTATTCTGAATGTTTTTCCACTAGCAAATATATTTAGCTCCTCTGAAGATGAGGTTGTTGTTGCCCCTGTT ........................................................................................................................................................................................................((((((....(((((.......(((((((.......)))))))..(((((((...(((...)))..))))))).........)))))..))))))....................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................TAGATCTGTGTGTCATCTGCATAAT........................................................................................................... | 25 | 2.00 | 0.00 | 2.00 | - | - | |
| ..........................................................................................................................................................................................................................TAGATCTGTGTGTCATCTGCATATAA.......................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | - | |
| ..............................................................................................................................................................................................................................TCTGTGTGTCATCTGCATAGAGATGAGC.................................................................................................... | 28 | 1.00 | 0.00 | - | 1.00 | - | |
| ........................................................................................................................................................................................................................TGTAGATCTGTGTGTCATCTAAAA.............................................................................................................. | 24 | 1.00 | 0.00 | 1.00 | - | - | |
| ..........................................................................................................................................................................................................................TAGATCTGTGTGTCATCTGCATAAAA.......................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | - | |
| .TGACACCGAGGTTGCGGGCCTGAGAAAAA................................................................................................................................................................................................................................................................................................................................ | 29 | 1.00 | 0.00 | 1.00 | - | - | |
| ..........................................................................................................................................................................................................................TAGATCTGTGTGTCATCTGCACC............................................................................................................. | 23 | 1.00 | 0.00 | 1.00 | - | - | |
| .................................................................................................................................................................................................................................................................................................................AATATATTTAGCTCCTCTGAAGATG.................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| .............TGCGGGCCTGAGAGACGGGAAGGAAA....................................................................................................................................................................................................................................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | - | |
| ........................................................................................................................................................................................................................TGTAGATCTGTGTGTAAA.................................................................................................................... | 18 | 1.00 | 0.00 | 1.00 | - | - | |
| ................GGGCCTGAGAGACGGGAAGGTAAA...................................................................................................................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | |
| ........................................................................................................................................................................................................................TGTAGATCTGTGTGTCATCTGAAAA............................................................................................................. | 25 | 1.00 | 0.00 | 1.00 | - | - | |
| ....................................................................................................................................................................................................................GAGATGTAGATCTGTGTGAAA..................................................................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | |
| ..............................................................................................................................................................................................................................TCTGTGTGTCATCTGCATAGAGATGTAGG................................................................................................... | 29 | 1.00 | 0.00 | 1.00 | - | - |
| ATGACACCGAGGTTGCGGGCCTGAGAGACGGGAAGGATGGTCGTACCATCCACGGTGATAGGGAAGTCAAGGAGAAGACAGGTCTTGGGAGGGAAGATGAGGAGCCCAGTTTTGGTCATGTTGACTTCTGGTGGCGGGCAGACATCCAGGTAGAGACATCTTGGAGGCAGGAGGAGATACGAGCCTGAAGGGAGGTGGAGAGGACAGGGGCAGAGATGTAGATCTGTGTGTCATCTGCATAGAGATGATAGTTGAAGCCGTGTTGCACTGTCAAAAGAGTTATTCTGAATGTTTTTCCACTAGCAAATATATTTAGCTCCTCTGAAGATGAGGTTGTTGTTGCCCCTGTT ........................................................................................................................................................................................................((((((....(((((.......(((((((.......)))))))..(((((((...(((...)))..))))))).........)))))..))))))....................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|