| (1) HEART | (1) KIDNEY | (2) OTHER |
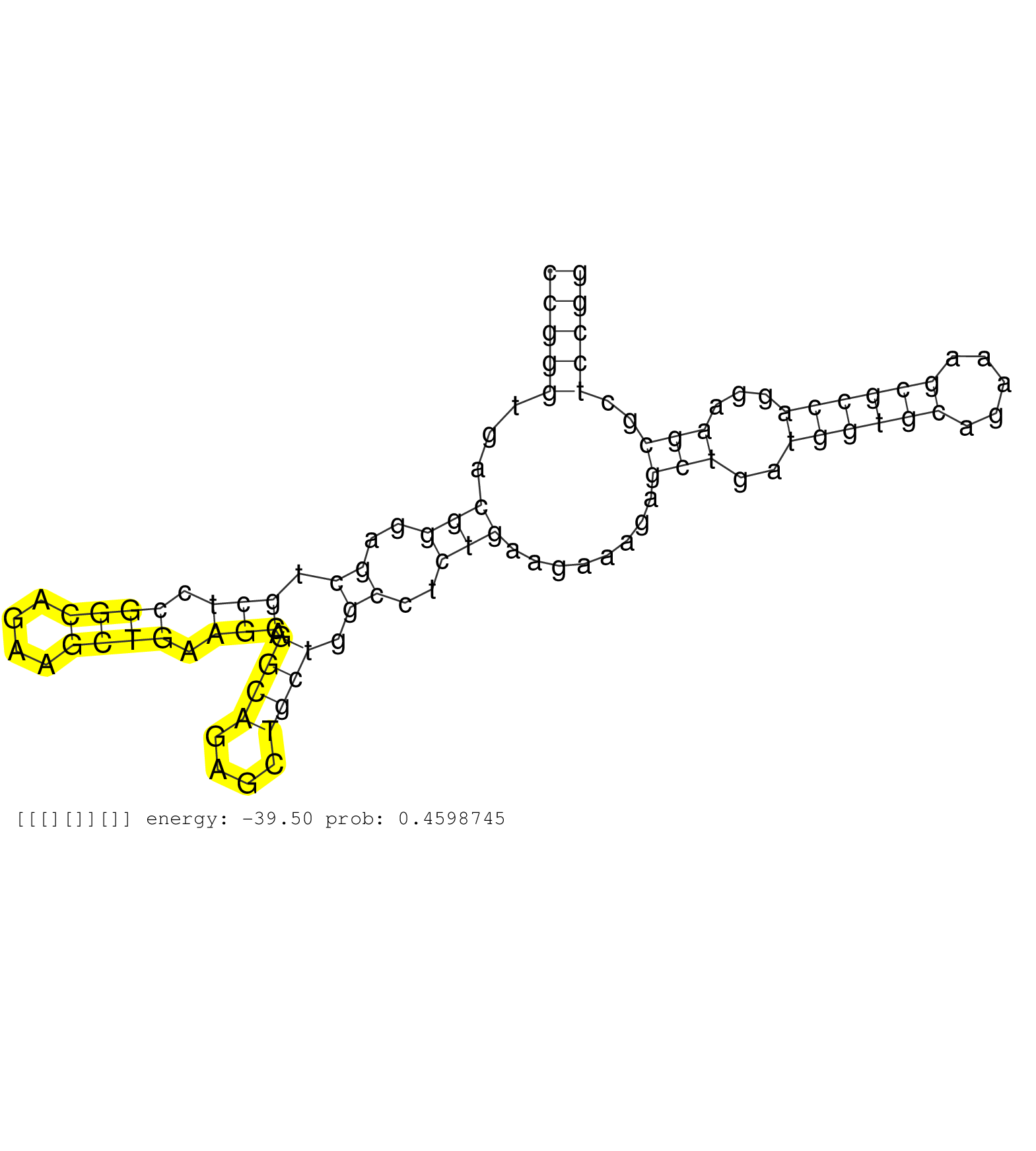
| CCCTTAATAAAGAGCTGATGGTGCAGAAACGCCAGGAAGCGCTCCAGGAACTTGNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNAGGAAAGGGGTGGGGCCATAACTGGGGGGGGCGTACATGGGGTCGCAGCGGGGGCGGAAGGGTGAGGATTGGGGTCGGGGGCAAGCCCGGTCCACCGACGGGGCTTGAGTCTCCCGGCCAGGGTATGTGGACCGGGTGACGGGAGCTGCTCCGGCAGAAGCTGAAGCAGGCAGAGCTGCTGGCCTCTGAAGAAAGAGCTGATGGTGCAGAAAGCGCCAGGAAGCGCTCCGGGAACAGGCCGCCCTGGAGCCCAAGCTGGACCTGCTCATAGACAAAACCAG ......................................................................................................................................................................................................................................(((((...(((..((.(((.((((....)))).))).((((....)))).))..)))........(((..((((((.....))))))...)))..))))).................................................. ......................................................................................................................................................................................................................................231................................................................................................330................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................................................................................................................................GCCAGGAAGCGCTCCGGGAACAGGC.......................................... | 25 | 1 | 2.00 | 2.00 | - | 2.00 | - | - |
| ...........................................................................................................................................................................................................................................................GGCAGAAGCTGAAGCAGGCAGAGCT........................................................................................................ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...........................................................................................................................................................................................................................................................GGCAGAAGCTGAAGCAGGCAGAGCTGCTG.................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .............................................................................................................................................................................................................................................................CAGAAGCTGAAGCAGGCAGAGCTGC...................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ........................................................................................................................................................................................................................................................TCCGGCAGAAGCTGAAGCAGGCAGAGC......................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ...............................................................................................................................................................................................................................................................GAAGCTGAAGCAGGCAGAGC......................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .................................................................................................................................................................................................................................................................................................................................................................GCTGGACCTGCTCATAGAC........ | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ............................................................................................................................................................................................................................................................................................CTGAAGAAAGAGCTGATGGTGCAGAAG..................................................................... | 27 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ...........................................................................................................................................................................................................................................................GGCAGAAGCTGAAGCAGGCAGAGCTGC...................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| CCCTTAATAAAGAGCTGATGGTGCAGAAACGCCAGGAAGCGCTCCAGGAACTTGNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNAGGAAAGGGGTGGGGCCATAACTGGGGGGGGCGTACATGGGGTCGCAGCGGGGGCGGAAGGGTGAGGATTGGGGTCGGGGGCAAGCCCGGTCCACCGACGGGGCTTGAGTCTCCCGGCCAGGGTATGTGGACCGGGTGACGGGAGCTGCTCCGGCAGAAGCTGAAGCAGGCAGAGCTGCTGGCCTCTGAAGAAAGAGCTGATGGTGCAGAAAGCGCCAGGAAGCGCTCCGGGAACAGGCCGCCCTGGAGCCCAAGCTGGACCTGCTCATAGACAAAACCAG ......................................................................................................................................................................................................................................(((((...(((..((.(((.((((....)))).))).((((....)))).))..)))........(((..((((((.....))))))...)))..))))).................................................. ......................................................................................................................................................................................................................................231................................................................................................330................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|