| (1) HEART | (1) KIDNEY | (1) OTHER |
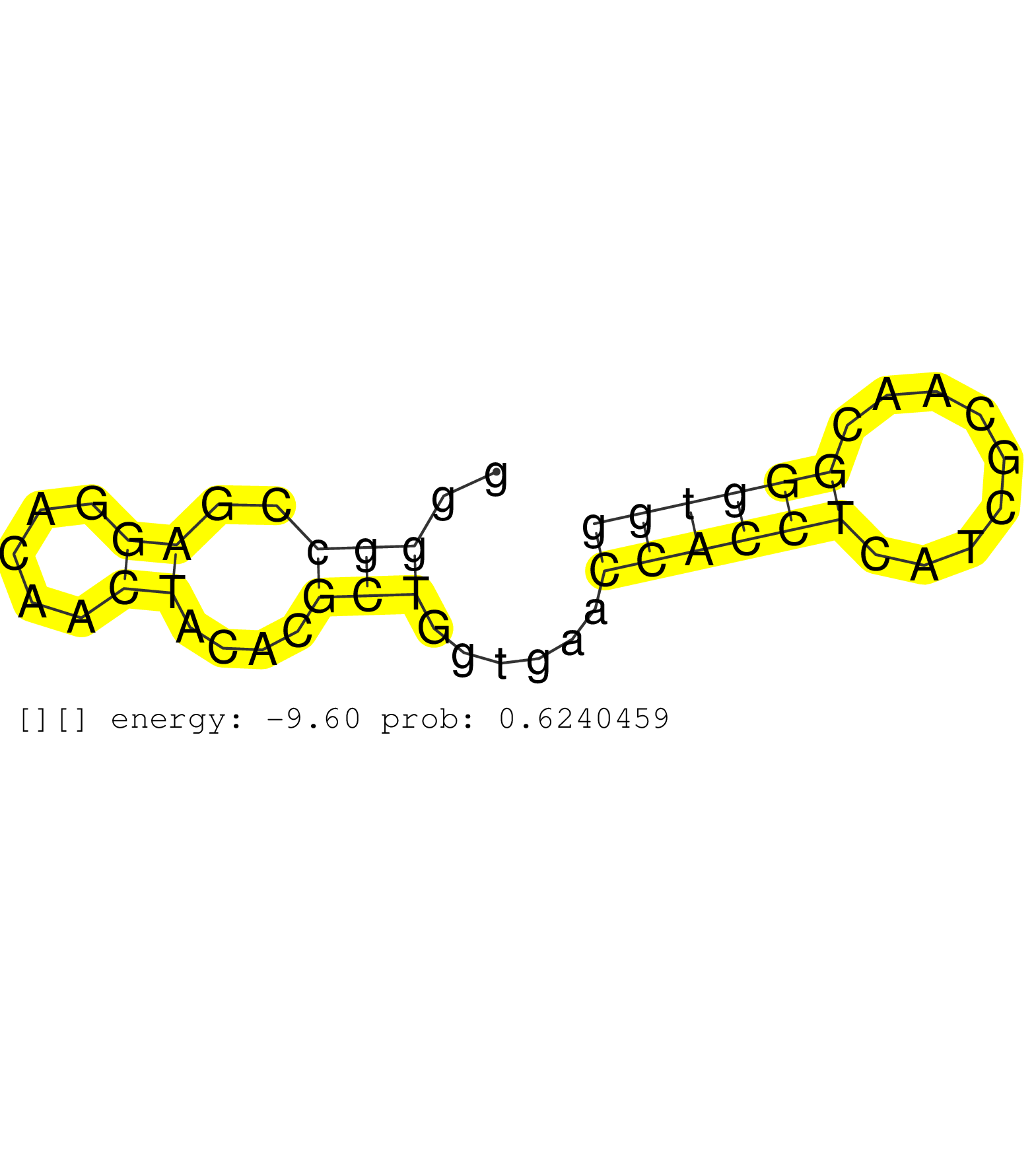
| GCTCTGGGTTGGACTCTCCTGAGGACTGCCTCTTGAGGGTCTCTCTCCCCTCCCCGGCCCCCCAGGTGGAACTCTACCAGTTCATGGCCAAGGACAACGTCCCTTTCCACAGCGTTGTCTTCCCCTGCTCACTGCTGGGGGCCGAGGACAACTACACGCTGGTGAACCACCTCATCGCAACGGGTGGGTGTGAGGCGGTGCCGGGGTGGGGGCGGCGAGGAGGGTGGTGGCTCAGGGGAGGGTGTCCGGGTGGGATGAGGTGAGGGGCCCTGACCCCCGGCCCCGCCGTGGTGCTCCCAGAATACCTGAACTACGAGGACGGGAAGTTCTCCAAGAGCCGGGGCGTGGGG ...........................................................................................................................................(((..((.....))....)))......((((((.........))))))................................................................................................................................................................... .........................................................................................................................................138..............................................187................................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|
| .........................................................................................AAGGACAACGTCCCTTTCCACAGCGTT.......................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ................................................................................................................................................AGGACAACTACACGC............................................................................................................................................................................................... | 15 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..............................................................................................................................................CGAGGACAACTACACGCTG............................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .....................................................................................GGCCAAGGACAACGT.......................................................................................................................................................................................................................................................... | 15 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...........................................................................................................................................................................................................................................................................................................GAATACCTGAACTACGAGGAC.............................. | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .................................................................................................................................................................GTGAACCACCTCATCGCAACGG....................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...............................................................................................................................................GAGGACAACTACACGCTGGTGAACC...................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .....................AGGACTGCCTCTTGAGG........................................................................................................................................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ......................................................................................................................................................................CCACCTCATCGCAACGG....................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .................................................................GTGGAACTCTACCAGTTCATGGCCAAGGACA.............................................................................................................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| .....................................................................................................................................................................................................................................................................................................................................GTTCTCCAAGAGCCGG......... | 16 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..................................................................................................................................................................................................................................................................................................................TGAACTACGAGGACGCA........................... | 17 | 1.00 | 0.00 | - | 1.00 | - | |
| ..................................................................................................................................................................................................................................................................................................................TGAACTACGAGGACGGGAAGT....................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...........................................................................................................................................................................................................................................................................................................................AGGACGGGAAGTTCTCCAAGAGCCGGGGC...... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ....................................................................................................................................................................................................................................................................................................GCTCCCAGAATACCTGAAC....................................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .......................................................................................................................................................................CACCTCATCGCAACGG....................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ..............................................................................................................................................................................................................................................................................................................TACCTGAACTACGAGGACGG............................ | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..................................................................................................................................................................................................................................................................................................................TGAACTACGAGGACGG............................ | 16 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| GCTCTGGGTTGGACTCTCCTGAGGACTGCCTCTTGAGGGTCTCTCTCCCCTCCCCGGCCCCCCAGGTGGAACTCTACCAGTTCATGGCCAAGGACAACGTCCCTTTCCACAGCGTTGTCTTCCCCTGCTCACTGCTGGGGGCCGAGGACAACTACACGCTGGTGAACCACCTCATCGCAACGGGTGGGTGTGAGGCGGTGCCGGGGTGGGGGCGGCGAGGAGGGTGGTGGCTCAGGGGAGGGTGTCCGGGTGGGATGAGGTGAGGGGCCCTGACCCCCGGCCCCGCCGTGGTGCTCCCAGAATACCTGAACTACGAGGACGGGAAGTTCTCCAAGAGCCGGGGCGTGGGG ...........................................................................................................................................(((..((.....))....)))......((((((.........))))))................................................................................................................................................................... .........................................................................................................................................138..............................................187................................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|
| .....................................................................................................AAGACAACGCTGTGGAAAGG..................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - |