| (1) OTHER |
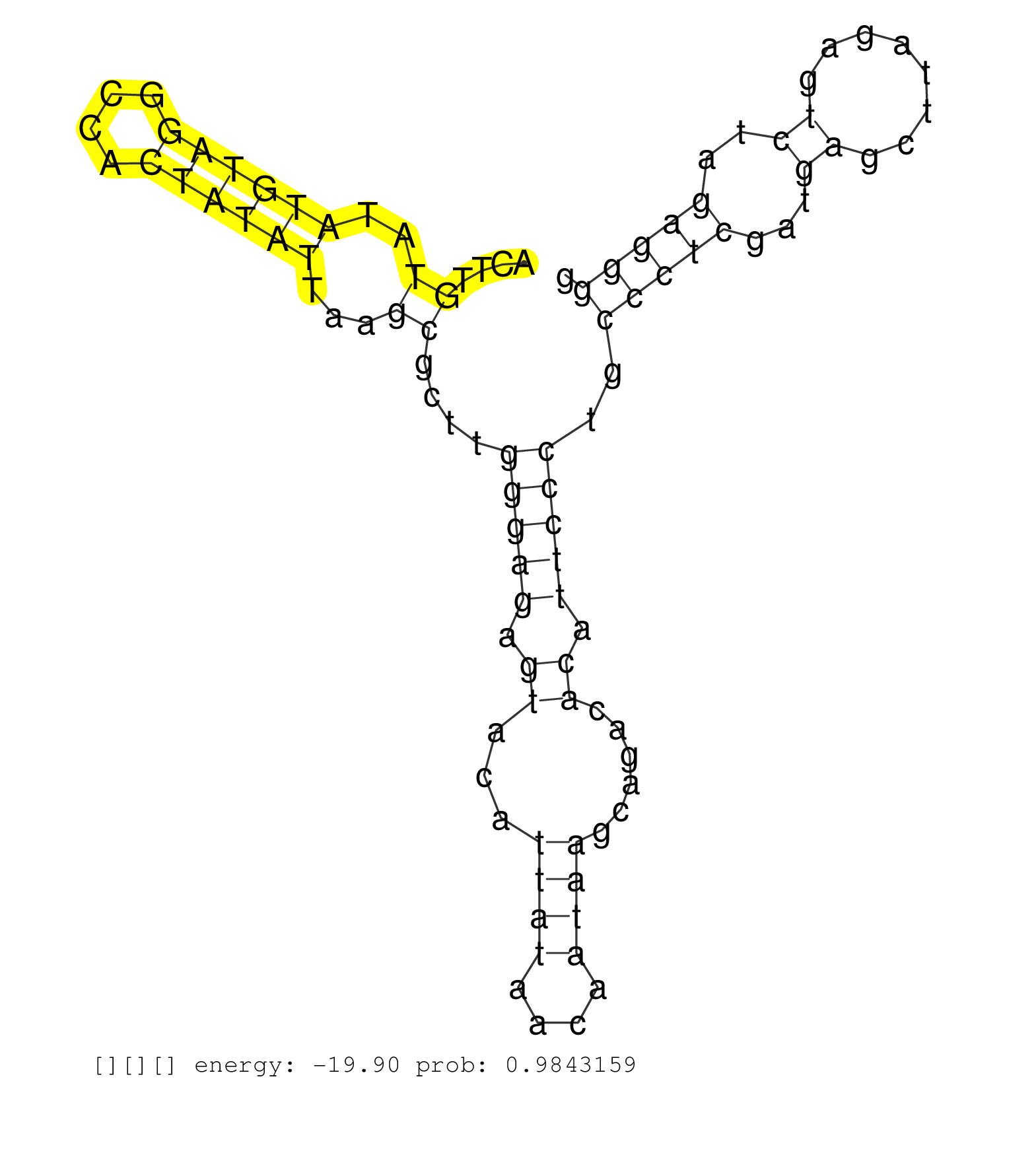
| GTTCCTCCATATTAGTCAAAAGGTGACAAGAATCAGGGATTGTCCTGTTTCTGAAGTGGATGGGAGTTTCTTGGATATCAACCGGTCTTGCCTGGCCCCCAAGAGGTTGATTACCTCAAAGACTGTGTCCCTGATGGTCAAAACACACAACGTTCATTCATTTATTCATCCATTCATTCTATCAATCGTGTTTATTGAGCACTTGTATATGTAGGCCACTATATTAAGCGCTTGGGAGAGTACATTATAACAATAAGCAGACACATTCCCTGCCCTCGATGAGCTTAGAGTCTAGAGGGGTTGTTGGTCACACTCCAGATTTCATGATCCTTTGTAAGCCCATATTTGGC ............................................................................................................................................................................................................((..((((((....))))))...))....(((((.((...((((....))))......)).)))))..(((((...((........))..)))))................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|
| ............................................................................................................................................................................................TGTTTATTGAGCACTTGTATATGTAGGC...................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 |
| ...................................................................................................................................................................................................TGAGCACTTGTATATGTAGGCCACTATATT............................................................................................................................. | 30 | 1 | 2.00 | 2.00 | 2.00 |
| ............................................................................................................................................................................................................................................................................................TTAGAGTCTAGAGGGGTTGTTGGTCATA...................................... | 28 | 2.00 | 0.00 | 2.00 | |
| .......................................TTGTCCTGTTTCTGAAGTGGATGGG.............................................................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 |
| ........................................................................................................................................................................................................ACTTGTATATGTAGGCCACTATATT............................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 |
| ...............................................................................................................................................................................................................................................................................................................................TTTCATGATCCTTTGTAAGCCCATATTCGGC | 31 | 1.00 | 0.00 | 1.00 | |
| ...................................................................................................................................................................................................TGAGCACTTGTATATGTAGGCCACTACT............................................................................................................................... | 28 | 1.00 | 0.00 | 1.00 | |
| ...........TTAGTCAAAAGGTGACAAGAATCAGGG........................................................................................................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 |
| ........................................................TGGATGGGAGTTTCTTGGATATCAACCGGT........................................................................................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | 1.00 |
| ............................................................................................................................................................................................TGTTTATTGAGCACTTGTATATGTAGGCC..................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 |
| .......................................................................................................................................................................................................................................................................................TGAGCTTAGAGTCTAGAGGGGTTGTTGGT.......................................... | 29 | 2 | 1.00 | 1.00 | 1.00 |
| ............................................................................................................................................................................................................................................................................................TTAGAGTCTAGAGGGGTTGTTGGTC......................................... | 25 | 2 | 0.50 | 0.50 | 0.50 |
| .......................................................................................................................................................................................................................................................................................TGAGCTTAGAGTCTAGAGGGGTTGTTG............................................ | 27 | 2 | 0.50 | 0.50 | 0.50 |
| .......................................................................................................................................................................................................................................................................................TGAGCTTAGAGTCTAGAGGGGTTGTTGG........................................... | 28 | 2 | 0.50 | 0.50 | 0.50 |
| ........................................................................................................................................................................................................................................TGGGAGAGTACATTATAACAATAAGCAGA......................................................................................... | 29 | 11 | 0.09 | 0.09 | 0.09 |
| GTTCCTCCATATTAGTCAAAAGGTGACAAGAATCAGGGATTGTCCTGTTTCTGAAGTGGATGGGAGTTTCTTGGATATCAACCGGTCTTGCCTGGCCCCCAAGAGGTTGATTACCTCAAAGACTGTGTCCCTGATGGTCAAAACACACAACGTTCATTCATTTATTCATCCATTCATTCTATCAATCGTGTTTATTGAGCACTTGTATATGTAGGCCACTATATTAAGCGCTTGGGAGAGTACATTATAACAATAAGCAGACACATTCCCTGCCCTCGATGAGCTTAGAGTCTAGAGGGGTTGTTGGTCACACTCCAGATTTCATGATCCTTTGTAAGCCCATATTTGGC ............................................................................................................................................................................................................((..((((((....))))))...))....(((((.((...((((....))))......)).)))))..(((((...((........))..)))))................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) |
|---|