| (1) BRAIN | (1) HEART | (1) KIDNEY | (2) OTHER |
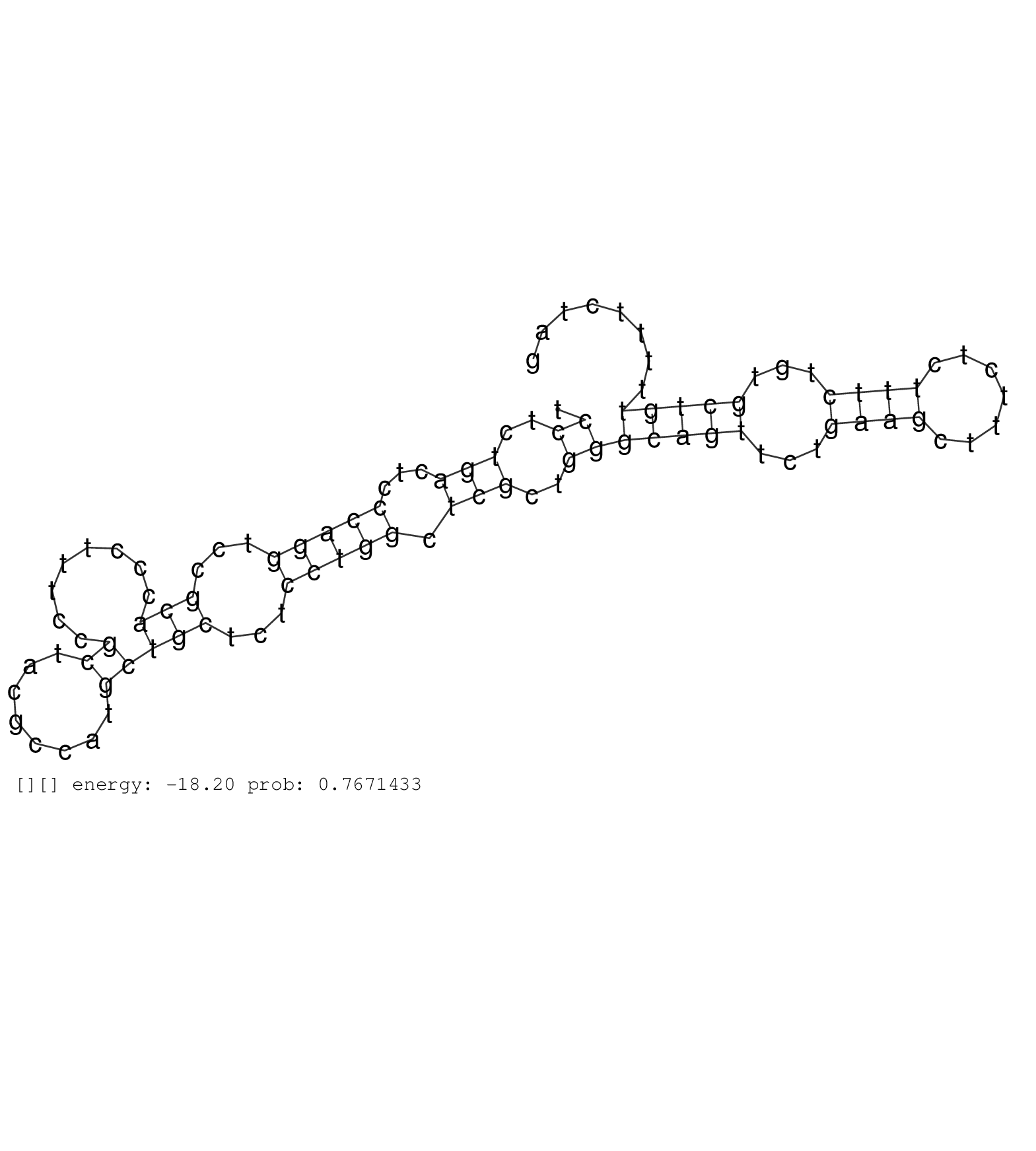
| AGTGCTTACGATGTGTCAGGCACTGTACTAAGCTCTGGGGGGCATACAAGCTAATCATGTTGGACCCAGTCCGTGTCCCACCTGGAGCTCACGGTCTTAATCCCCATTTTACAGATGAGGTAACCTAGGCCCAGAGAAGTGAAATGACTTGCCGAGGGTCACACAGCAGACAAGCTTCGGAGCCGGGGTTAGAACCCAGGTCCTCTGACTCCCAGGTCCGCACCCTTTCCGCTACGCCATGCTGCTCTCCTGGCTCGCTGGGCAGTTCTGAAGCTTTCTCTTTCTGTGCTGTTTTTCTAGGGGTCACAACGGAGTGCGGTCCTGCATCAACTGTTTCAGCTCGGACGTGA .........................................................................................................................................................................................................((..(((...(((((...(((........((........)))))...))))).)))..))(((((...((((.......))))...))))).......................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553592(SRX182798) source: Brain. (Brain) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|---|
| .........................................................................TGTCCCACCTGGAGCTCACGGTA.............................................................................................................................................................................................................................................................. | 23 | 9.00 | 0.00 | 7.00 | 1.00 | 1.00 | - | - | |
| .........................................................................TGTCCCACCTGGAGCTCACGGTAT............................................................................................................................................................................................................................................................. | 24 | 8.00 | 0.00 | 4.00 | 3.00 | - | 1.00 | - | |
| .........................................................................TGTCCCACCTGGAGCTCACGGTAA............................................................................................................................................................................................................................................................. | 24 | 5.00 | 0.00 | 3.00 | - | 2.00 | - | - | |
| .........................................................................TGTCCCACCTGGAGCTCACGGA............................................................................................................................................................................................................................................................... | 22 | 3.00 | 0.00 | - | 2.00 | 1.00 | - | - | |
| .........................................................................TGTCCCACCTGGAGCTCACGGAAT............................................................................................................................................................................................................................................................. | 24 | 3.00 | 0.00 | 2.00 | 1.00 | - | - | - | |
| .........................................................................TGTCCCACCTGGAGCTCACGGTAAT............................................................................................................................................................................................................................................................ | 25 | 3.00 | 0.00 | 1.00 | 1.00 | 1.00 | - | - | |
| .....................................................................................................................................................................................................................................................................................................................................ATCAACTGTTTCAGCTCGGAAGCC. | 24 | 1.00 | 0.00 | - | - | - | - | 1.00 | |
| .....................................................................................................................................................................................................................................................................................................................................ATCAACTGTTTCAGCTCGGACG... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .........................................................................TGTCCCACCTGGAGCTCACGGAT.............................................................................................................................................................................................................................................................. | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| .........................................................................TGTCCCACCTGGAGCTCACGGTAAA............................................................................................................................................................................................................................................................ | 25 | 1.00 | 0.00 | - | 1.00 | - | - | - | |
| .........................................................................TGTCCCACCTGGAGCTCACGGTAAC............................................................................................................................................................................................................................................................ | 25 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| .........................................................................TGTCCCACCTGGAGCTCACGGATT............................................................................................................................................................................................................................................................. | 24 | 1.00 | 0.00 | - | 1.00 | - | - | - | |
| ..................................................................................................................................................................................................................................................................................................................................TGCATCAACTGTTTCAGCTCGGACGC.. | 26 | 1.00 | 0.00 | - | - | - | 1.00 | - |
| AGTGCTTACGATGTGTCAGGCACTGTACTAAGCTCTGGGGGGCATACAAGCTAATCATGTTGGACCCAGTCCGTGTCCCACCTGGAGCTCACGGTCTTAATCCCCATTTTACAGATGAGGTAACCTAGGCCCAGAGAAGTGAAATGACTTGCCGAGGGTCACACAGCAGACAAGCTTCGGAGCCGGGGTTAGAACCCAGGTCCTCTGACTCCCAGGTCCGCACCCTTTCCGCTACGCCATGCTGCTCTCCTGGCTCGCTGGGCAGTTCTGAAGCTTTCTCTTTCTGTGCTGTTTTTCTAGGGGTCACAACGGAGTGCGGTCCTGCATCAACTGTTTCAGCTCGGACGTGA .........................................................................................................................................................................................................((..(((...(((((...(((........((........)))))...))))).)))..))(((((...((((.......))))...))))).......................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553592(SRX182798) source: Brain. (Brain) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................................................................................TGACCCCTAGAAAAACAGCACAGAAAGAGA............................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - |
| ....................................................................................................................................................................................atgTGGGTTCTAACCCCGGTA..................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | 1.00 | - | |
| ...................................................................................................................................................gcaCTGTGTGACCCTCGGCACG..................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - |