| (1) HEART | (1) KIDNEY | (2) OTHER |
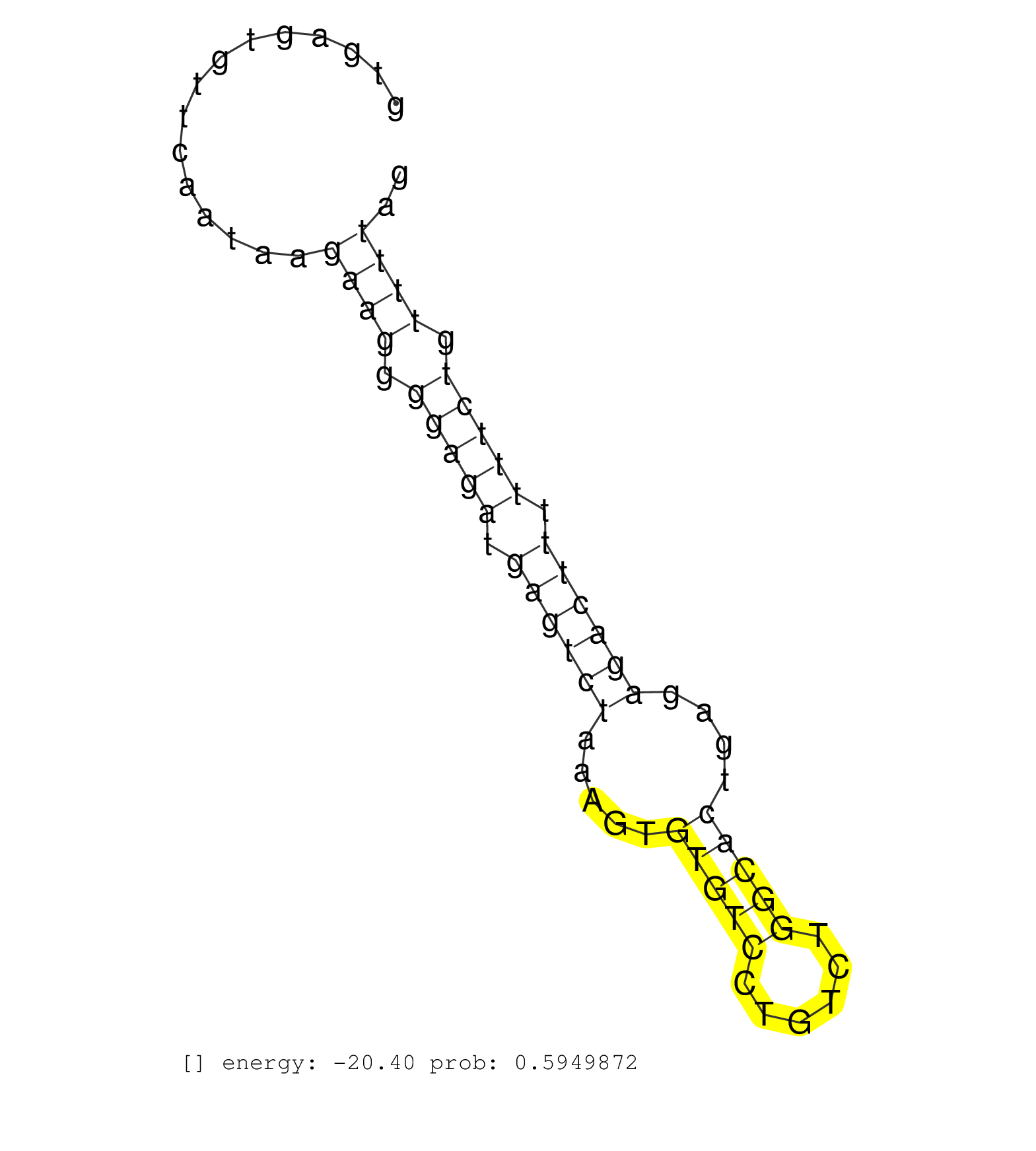
| TTGCAGGAGGAAGGGGTGGACTTGATGAACAGAGAAACAGCACATGAAAGGTGAGTGTTCAATAAGAAGGGGAGATGAGTCTAAAGTGTGTCCTGTCTGGCACTGAGAGACTTTTTTCTGTTTTAGGGAAGTGCAAACAGCAATGCAGATTAGTCAGTCATGGGAGGAGAGCCTAA .................................................................((((.(((((.((((((.....(((((......)))))....)))))).))))).)))).................................................... ..................................................51.........................................................................126................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|
| .......................GATGAACAGAGAAACAGCACATGAAAGGGA........................................................................................................................... | 30 | 2.00 | 0.00 | - | 2.00 | - | - | |
| ....................................................................................AGTGTGTCCTGTCTGGC........................................................................... | 17 | 1 | 2.00 | 2.00 | - | - | 2.00 | - |
| ..............................................................................................GTCTGGCACTGAGAGAC................................................................. | 17 | 2 | 1.50 | 1.50 | 1.50 | - | - | - |
| ...................................................................................................................................TGCAAACAGCAATGCAGATTAGTCAGTCATGGGA........... | 34 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .............GGGTGGACTTGATGAACAGA............................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ........GGAAGGGGTGGACTTGATGAACAGAGAAACA......................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..............................................................................................GTCTGGCACTGAGAGA.................................................................. | 16 | 2 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..............GGTGGACTTGATGAAC.................................................................................................................................................. | 16 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .....................TTGATGAACAGAGAAACAGCA...................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..................GACTTGATGAACAGAGAAACAGCACATG.................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ................TGGACTTGATGAACAGAGAAACAGC....................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ...............GTGGACTTGATGAACAGAG.............................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .........GAAGGGGTGGACTTGATGAACAGAGAAAC.......................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..................GACTTGATGAACAGAGAAAC.......................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...............GTGGACTTGATGAACAGAGAAACAGC....................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| .........GAAGGGGTGGACTTGATGAACAGAG.............................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...............................................................................................TCTGGCACTGAGAGAC................................................................. | 16 | 2 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..................GACTTGATGAACAGAGAAACAGCA...................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .........................TGAACAGAGAAACAGCACATGAAAGGGAA.......................................................................................................................... | 29 | 1.00 | 0.00 | - | - | 1.00 | - | |
| .......AGGAAGGGGTGGACTTGATGAACAGAG.............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ..................GACTTGATGAACAGAGAAACAGCACA.................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .........GAAGGGGTGGACTTGATGAACAGAGAAACA......................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .......................................................................................................................................AACAGCAATGCAGATTA........................ | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .......................................................................................................................................AACAGCAATGCAGATTAGTCAGTCATGGGAG.......... | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .......................GATGAACAGAGAAACAGC....................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ......................TGATGAACAGAGAAACAGCACATGAAAGG............................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ......GAGGAAGGGGTGGACTTGATGAAC.................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...............GTGGACTTGATGAACAGAGAAACAGCACATG.................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| .........GAAGGGGTGGACTTGATGAACAGAGAA............................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .........GAAGGGGTGGACTTGATGAACA................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .......................GATGAACAGAGAAACAGCA...................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ......GAGGAAGGGGTGGACTTGA....................................................................................................................................................... | 19 | 2 | 0.50 | 0.50 | 0.50 | - | - | - |
| ......................TGATGAACAGAGAAACA......................................................................................................................................... | 17 | 3 | 0.33 | 0.33 | 0.33 | - | - | - |
| .........................TGAACAGAGAAACAGCA...................................................................................................................................... | 17 | 5 | 0.20 | 0.20 | - | - | - | 0.20 |
| .............................................................................................TGTCTGGCACTGAGA.................................................................... | 15 | 8 | 0.12 | 0.12 | 0.12 | - | - | - |
| ................................................................................................CTGGCACTGAGAGAC................................................................. | 15 | 9 | 0.11 | 0.11 | 0.11 | - | - | - |
| ...........................AACAGAGAAACAGCAC..................................................................................................................................... | 16 | 11 | 0.09 | 0.09 | 0.09 | - | - | - |
| TTGCAGGAGGAAGGGGTGGACTTGATGAACAGAGAAACAGCACATGAAAGGTGAGTGTTCAATAAGAAGGGGAGATGAGTCTAAAGTGTGTCCTGTCTGGCACTGAGAGACTTTTTTCTGTTTTAGGGAAGTGCAAACAGCAATGCAGATTAGTCAGTCATGGGAGGAGAGCCTAA .................................................................((((.(((((.((((((.....(((((......)))))....)))))).))))).)))).................................................... ..................................................51.........................................................................126................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|