| (1) HEART | (1) KIDNEY |
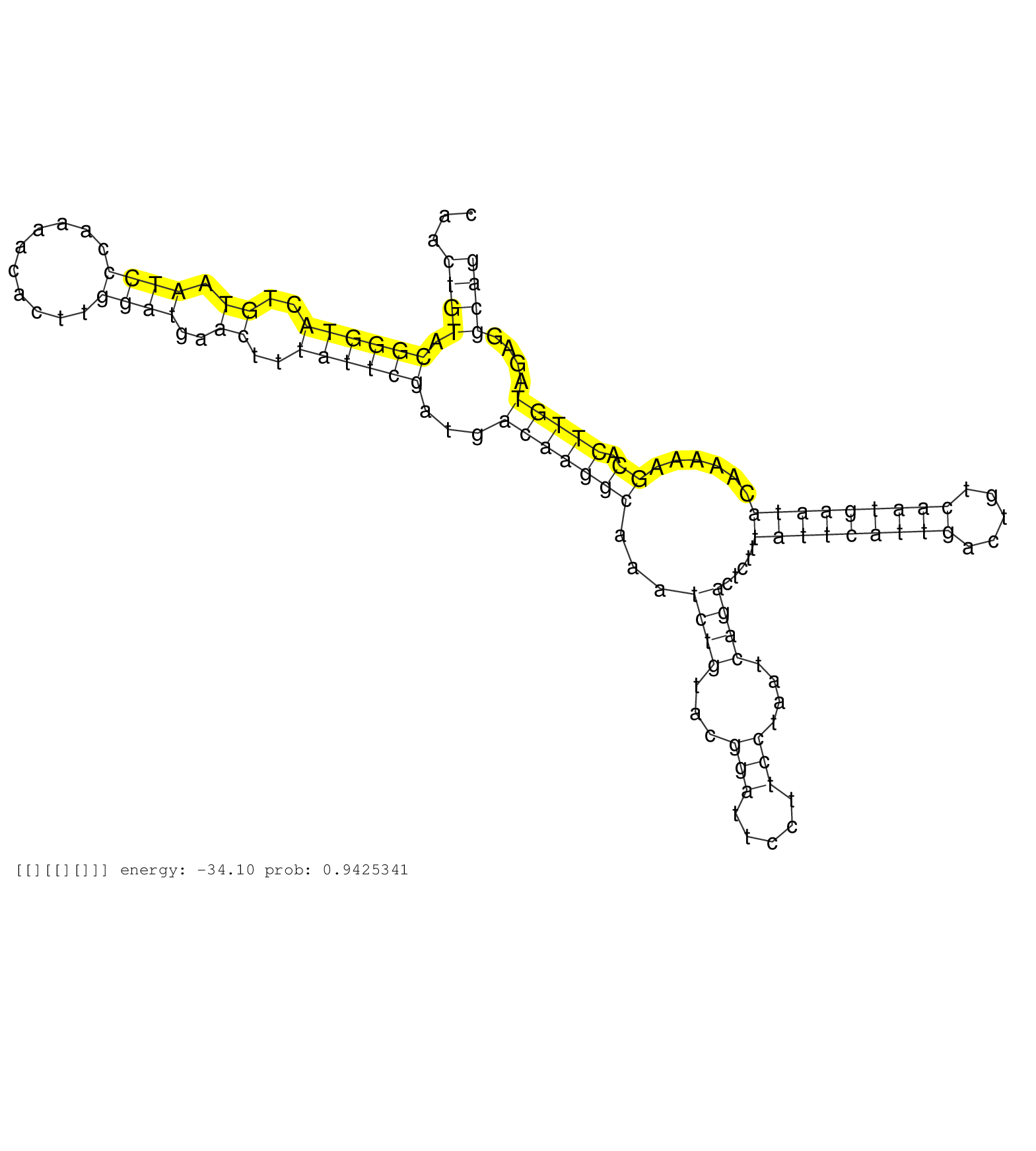
| TCGAGAGCCCAGCGGATGCCAAGGATGCTGCCAGAGACATGAATGGAAAGGTGTGTGGTTTTGTTTCCTTGTCTTGTCTTTTATCCCCCCCTCCCCCCCCCAATGGTACCTACACTCCTCCGGGAAACGTCGTGCAGGCTTCTCGTCCCAACTGTACGGGTACTGTAATCCCAAAACACTTGGATGAACTTTATTCGATGACAAGGCAAATCTGTACGGATTCCTTCCTAATCAGACTCTTTATTCATTGACTGTCAATGAATACAAAAAGCACTTGTAGAGGCAGATAGCCTTTTCGAAGGCTTAAATGGGCTTGTATCTTGTACATTTACATTTTGTGCTCTGTATTA .......................................................................................................................................................((((.((((((..((.((((..........))))..))..))))))...(((((((...((((...(((.....)))....)))).....(((((((((.....)))))))))......)).)))))....))))................................................................ ....................................................................................................................................................149......................................................................................................................................286.............................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................AAGGCAAATCTGTACG.................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | 1.00 | - |
| ..........AGCGGATGCCAAGGATGCTGCCAGA........................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 |
| ....................AAGGATGCTGCCAGAGACATGAATGGA............................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - |
| .........................................................................................................................................................................................GAACTTTATTCGATGACAAGGCAAATCTGTAC..................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | 1.00 |
| .........................................................................................................................................................GTACGGGTACTGTAATC.................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | 1.00 | - |
| ........................................................................................................................................................................................................................................................................CAAAAAGCACTTGTAGAG.................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - |
| .......................................................................................................................................................................................................................................................................ACAAAAAGCACTTGTAGAGGCAGATAG............................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 |
| .......................................................................................................................................................................................ATGAACTTTATTCGATGACAAGGC............................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 |
| ....................AAGGATGCTGCCAGAGACAT...................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - |
| ....................................................................................................................................................................................TGGATGAACTTTATTCGATGACA................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - |
| ....................AAGGATGCTGCCAGA........................................................................................................................................................................................................................................................................................................................... | 15 | 5 | 0.20 | 0.20 | 0.20 | - |
| TCGAGAGCCCAGCGGATGCCAAGGATGCTGCCAGAGACATGAATGGAAAGGTGTGTGGTTTTGTTTCCTTGTCTTGTCTTTTATCCCCCCCTCCCCCCCCCAATGGTACCTACACTCCTCCGGGAAACGTCGTGCAGGCTTCTCGTCCCAACTGTACGGGTACTGTAATCCCAAAACACTTGGATGAACTTTATTCGATGACAAGGCAAATCTGTACGGATTCCTTCCTAATCAGACTCTTTATTCATTGACTGTCAATGAATACAAAAAGCACTTGTAGAGGCAGATAGCCTTTTCGAAGGCTTAAATGGGCTTGTATCTTGTACATTTACATTTTGTGCTCTGTATTA .......................................................................................................................................................((((.((((((..((.((((..........))))..))..))))))...(((((((...((((...(((.....)))....)))).....(((((((((.....)))))))))......)).)))))....))))................................................................ ....................................................................................................................................................149......................................................................................................................................286.............................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|