| (1) BRAIN | (1) HEART | (1) KIDNEY | (2) OTHER |
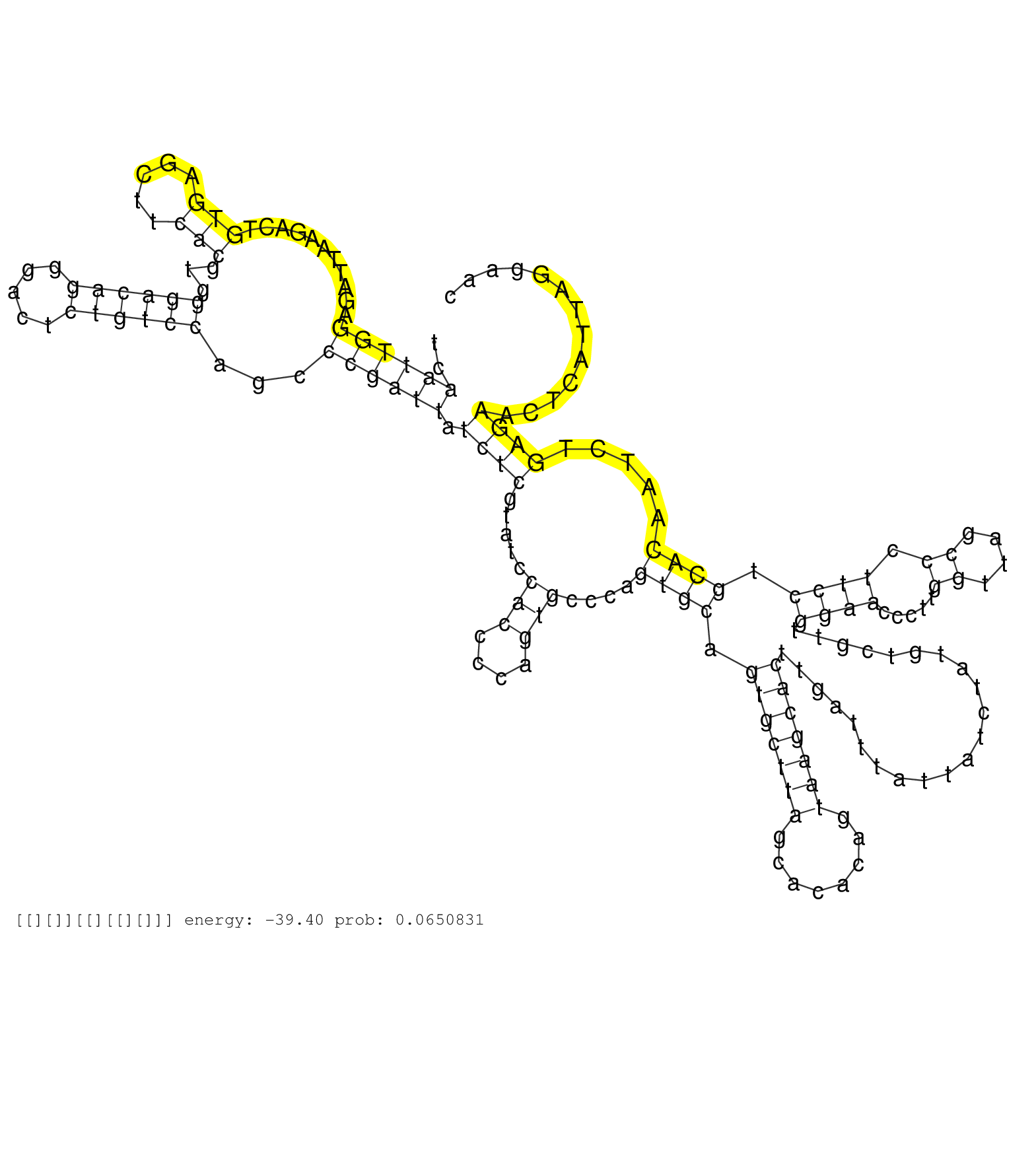
| GTGATCTGGGTTCTAATTCTGACTCTGCCACTTGTCTGCCGAGTGACCTTGGACAATTCCCTTAACTTCTTTAGGCCTTAGCCACCTCATATGTCAATTGGAGATTAAGACTGTGAGCTTCACGTGGGACAGGGACTCTGTCCAGCCCGATTATCTCGTATCCACCCCAGTGCCCAGTGCAGTGCTTAGCACACAGTAAGCACTTGATTTATTATCTATGTCGTTGGAACCCTTGGTTAGCCCTTCCTGCACAATCTGAGAACTCATTAGGAACATTCAGATCTGTATTTTCTCTTGCAGCGGCCGGGGCTTTGTCCAAGCTAACGCACGGATTGAAAGACGAGTCATTG ...............................................................................................((((((...........(((.....)))...((((((.....))))))...)))))).((((.....(((....)))....((((.(((((((........)))))))......................((((.....((....)).)))).)))).....))))......................................................................................... .............................................................................................94..................................................................................................................................................................................274.......................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553592(SRX182798) source: Brain. (Brain) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................TGGAGATTAAGACTGTTAAA........................................................................................................................................................................................................................................ | 20 | 37.00 | 0.00 | 37.00 | - | - | - | - | |
| ..................................................................................................TGGAGATTAAGACTGTGCGAA....................................................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| ............................................................................................................................................................................................................................................................................................................................CAAGCTAACGCACGGATTGAAAGACGAGTCATT. | 33 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ..................................................................................................TGGAGATTAAGACTGTGCAAT....................................................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| ........................................................................................................TTAAGACTGTGAGCTTCACGTTGGA............................................................................................................................................................................................................................. | 25 | 1.00 | 0.00 | - | - | 1.00 | - | - | |
| ......................................................................................................................................................................................................................................................................................................................................CACGGATTGAAAGACGAGTCAT.. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| .........................................................................................................................................................................................................................................................CACAATCTGAGAACTCATTAG................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 |
| ..................................................................................................TGGAGATTAAGACTGTTATT........................................................................................................................................................................................................................................ | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| .................................................................................................................................................................................................................................................................GAGAACTCATTAGGAACATTCAGATCTG................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - |
| ....................................................................................................................................................................................................................................................................................................................................CGCACGGATTGAAAGACGCGTC.... | 22 | 1.00 | 0.00 | - | - | - | 1.00 | - | |
| ..................................................................................................TGGAGATTAAGACTGTGCAA........................................................................................................................................................................................................................................ | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| ....................................................................................................................................................................................................................................................................................................................................CGCACGGATTGAAAGACGAGTCATTAA | 27 | 1.00 | 0.00 | - | - | 1.00 | - | - | |
| .......................................................................................................................................................................................................................................................................................................................................ACGGATTGAAAGACGA....... | 16 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................................................................................CCTGCACAATCTGAG.......................................................................................... | 15 | 4 | 0.25 | 0.25 | - | - | 0.25 | - | - |
| ..................................................................................................TGGAGATTAAGACTGTGAGCTTCACGT................................................................................................................................................................................................................................. | 27 | 10 | 0.10 | 0.10 | 0.10 | - | - | - | - |
| ..........................................................................................................AAGACTGTGAGCTTCACGTGGGACAG.......................................................................................................................................................................................................................... | 26 | 17 | 0.06 | 0.06 | - | 0.06 | - | - | - |
| GTGATCTGGGTTCTAATTCTGACTCTGCCACTTGTCTGCCGAGTGACCTTGGACAATTCCCTTAACTTCTTTAGGCCTTAGCCACCTCATATGTCAATTGGAGATTAAGACTGTGAGCTTCACGTGGGACAGGGACTCTGTCCAGCCCGATTATCTCGTATCCACCCCAGTGCCCAGTGCAGTGCTTAGCACACAGTAAGCACTTGATTTATTATCTATGTCGTTGGAACCCTTGGTTAGCCCTTCCTGCACAATCTGAGAACTCATTAGGAACATTCAGATCTGTATTTTCTCTTGCAGCGGCCGGGGCTTTGTCCAAGCTAACGCACGGATTGAAAGACGAGTCATTG ...............................................................................................((((((...........(((.....)))...((((((.....))))))...)))))).((((.....(((....)))....((((.(((((((........)))))))......................((((.....((....)).)))).)))).....))))......................................................................................... .............................................................................................94..................................................................................................................................................................................274.......................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553592(SRX182798) source: Brain. (Brain) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|---|
| ........................................................aaaAAGAAGTTAAGGAAA.................................................................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - |