| (1) HEART | (1) KIDNEY | (1) OTHER |
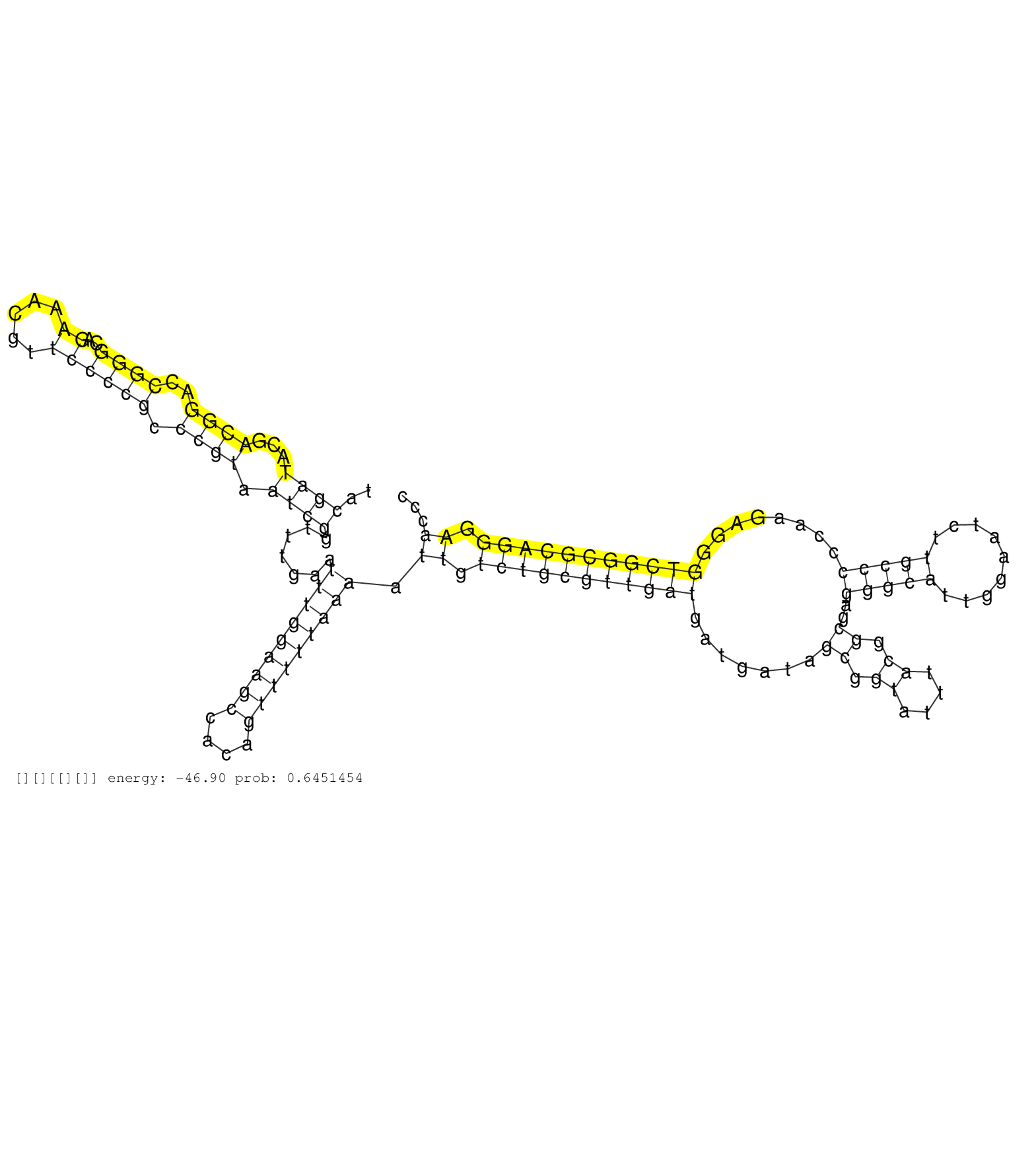
| GCACGGTTCCTATCGGTTTTTTTTTTTTGCGTCCTTGCTCTGTGCCGAGCATCCGGGGAGAGTACGATACGACGGACCGGGCAGAAACGTTCCCCGCCCGTAATCGGTTTGAATTTGGAAGCCACAGTTTTTAAAATTGTCTGCGTTGATGATGATAGCGGTATTTACGGCGAGGGCATTGGAATCTTGCCCCCAAGAGGGTCGGCGCAGGGAACCCTGTAGAGCCCGTCCTCCGTCCCCTCTGGCTCGCCCCTAGGCCGTGGGGGAAGGGCTGGCCCCAAGTGAGCCCGTGTCTCCCGGTCCGAGGGAGACTGCACCGCAAGGGCCCTTCTGCCTGGAAACCCCTCCCG ................................................................((((...((((..((((..((.....)))))).)))).)))).......(((((((((....))))))))).((.(((((((((((.......((.((....)).))..(((((.........)))))........))))))))))).))........................................................................................................................................ ..............................................................63........................................................................................................................................................217................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| ..........................................................AGAGTACGATACGACGGACCGGGCAG.......................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ............................................................AGTACGATACGACGGACCGGGCAGAAACG..................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...................................................................TACGACGGACCGGGCAGAAACGTTCCCC............................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..................................................................................................................................................TGATGATGATAGCGGTATTTACGGCGAGT............................................................................................................................................................................... | 29 | 1.00 | 0.00 | 1.00 | - | - | |
| ................................................................CGATACGACGGACCGGGCAGAAACGTTCC................................................................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .....................................................................................................................................................................................................AGGGTCGGCGCAGGGAACCCTGTAGA............................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ...................................................................TACGACGGACCGGGCAGAAAC...................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ....................................................................................................................................................................................................GAGGGTCGGCGCAGGCC......................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | 1.00 | |
| ...................................................................TACGACGGACCGGGCAGAAACGTTCC................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| GCACGGTTCCTATCGGTTTTTTTTTTTTGCGTCCTTGCTCTGTGCCGAGCATCCGGGGAGAGTACGATACGACGGACCGGGCAGAAACGTTCCCCGCCCGTAATCGGTTTGAATTTGGAAGCCACAGTTTTTAAAATTGTCTGCGTTGATGATGATAGCGGTATTTACGGCGAGGGCATTGGAATCTTGCCCCCAAGAGGGTCGGCGCAGGGAACCCTGTAGAGCCCGTCCTCCGTCCCCTCTGGCTCGCCCCTAGGCCGTGGGGGAAGGGCTGGCCCCAAGTGAGCCCGTGTCTCCCGGTCCGAGGGAGACTGCACCGCAAGGGCCCTTCTGCCTGGAAACCCCTCCCG ................................................................((((...((((..((((..((.....)))))).)))).)))).......(((((((((....))))))))).((.(((((((((((.......((.((....)).))..(((((.........)))))........))))))))))).))........................................................................................................................................ ..............................................................63........................................................................................................................................................217................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| ........gctAAAAAAACCGATTCG.................................................................................................................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | 1.00 |