| (1) HEART | (1) KIDNEY | (1) OTHER |
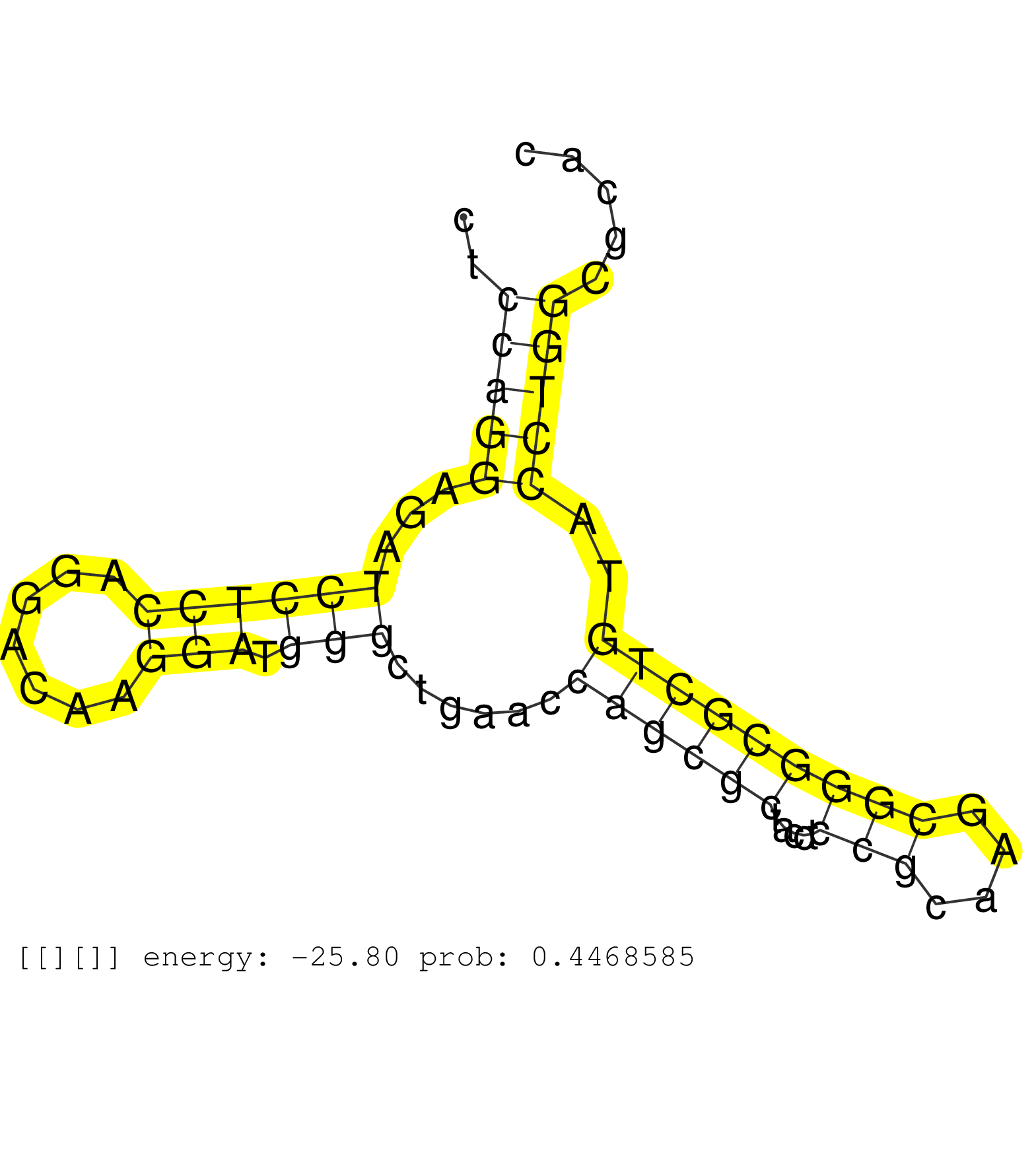
| TCTACCAGCTGGAAGCTGGCCCACTGGGCCCCGGTCCTGGGGGAGGAAACGTGAAGCCCTGAAGCCCCTTTGCCCCCGGTCTTCCTCCTCCAGGAGATCCTCCAGGACAAGGATGGGCTGAACCAGCGCTACCTCCGCAAGCGGGCGCTGTACCTGGCGCACCTCGCGCACCACCTGGCACGGGACCCCCTCTTCGGCAGCGTCCGCTTCACCTACATGAACGGCTGCCACCTCAAGCCCCTGCTGCTGCTGCGGCCCCAGGGTGCGGTGGTGGCGGGGGTGGGAGGCGACGGGGGAGGGTGGATGCCGAAGANNNNNNNNNNAATGTTTAATATGAACAAATAAGTAAT .........................................................................................(((((...((((((.......))).)))......((((((.....(((....)))))))))..)))))................................................................................................................................................................................................. .......................................................................................88........................................................................162.......................................................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................AGCGGGCGCTGTACCTGGC................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ............................................................................................GGAGATCCTCCAGGACAAGGAT............................................................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ............................................................................................GGAGATCCTCCAGGACAAGGATGGGCTGAACC.................................................................................................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .........................................................................................................GACAAGGATGGGCTGAACCAGC............................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ................................................................................................................ATGGGCTGAACCAGCGCTACC......................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ........................................................................................................GGACAAGGATGGGCTGAACCA................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ............................................................................................................AAGGATGGGCTGAACCA................................................................................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...........................................................................................AGGAGATCCTCCAGGACAAGGA............................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .....................................................................................................................................TCCGCAAGCGGGCGCTGTACCTG.................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ............................................................................................................AAGGATGGGCTGAACC.................................................................................................................................................................................................................................. | 16 | 2 | 0.50 | 0.50 | 0.50 | - | - |
| .............................................................................................................AGGATGGGCTGAACCA................................................................................................................................................................................................................................. | 16 | 2 | 0.50 | 0.50 | 0.50 | - | - |
| ...........................................................................................................................................AGCGGGCGCTGTACC.................................................................................................................................................................................................... | 15 | 3 | 0.33 | 0.33 | - | 0.33 | - |
| ......................................................................................................CAGGACAAGGATGGGC........................................................................................................................................................................................................................................ | 16 | 6 | 0.17 | 0.17 | 0.17 | - | - |
| .......................................................................................................AGGACAAGGATGGGC........................................................................................................................................................................................................................................ | 15 | 15 | 0.07 | 0.07 | 0.07 | - | - |
| TCTACCAGCTGGAAGCTGGCCCACTGGGCCCCGGTCCTGGGGGAGGAAACGTGAAGCCCTGAAGCCCCTTTGCCCCCGGTCTTCCTCCTCCAGGAGATCCTCCAGGACAAGGATGGGCTGAACCAGCGCTACCTCCGCAAGCGGGCGCTGTACCTGGCGCACCTCGCGCACCACCTGGCACGGGACCCCCTCTTCGGCAGCGTCCGCTTCACCTACATGAACGGCTGCCACCTCAAGCCCCTGCTGCTGCTGCGGCCCCAGGGTGCGGTGGTGGCGGGGGTGGGAGGCGACGGGGGAGGGTGGATGCCGAAGANNNNNNNNNNAATGTTTAATATGAACAAATAAGTAAT .........................................................................................(((((...((((((.......))).)))......((((((.....(((....)))))))))..)))))................................................................................................................................................................................................. .......................................................................................88........................................................................162.......................................................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................................................GGGGCCGCAGCAGCA........................................................................................... | 15 | 6 | 0.17 | 0.17 | 0.17 | - | - |