| (1) HEART | (2) OTHER |
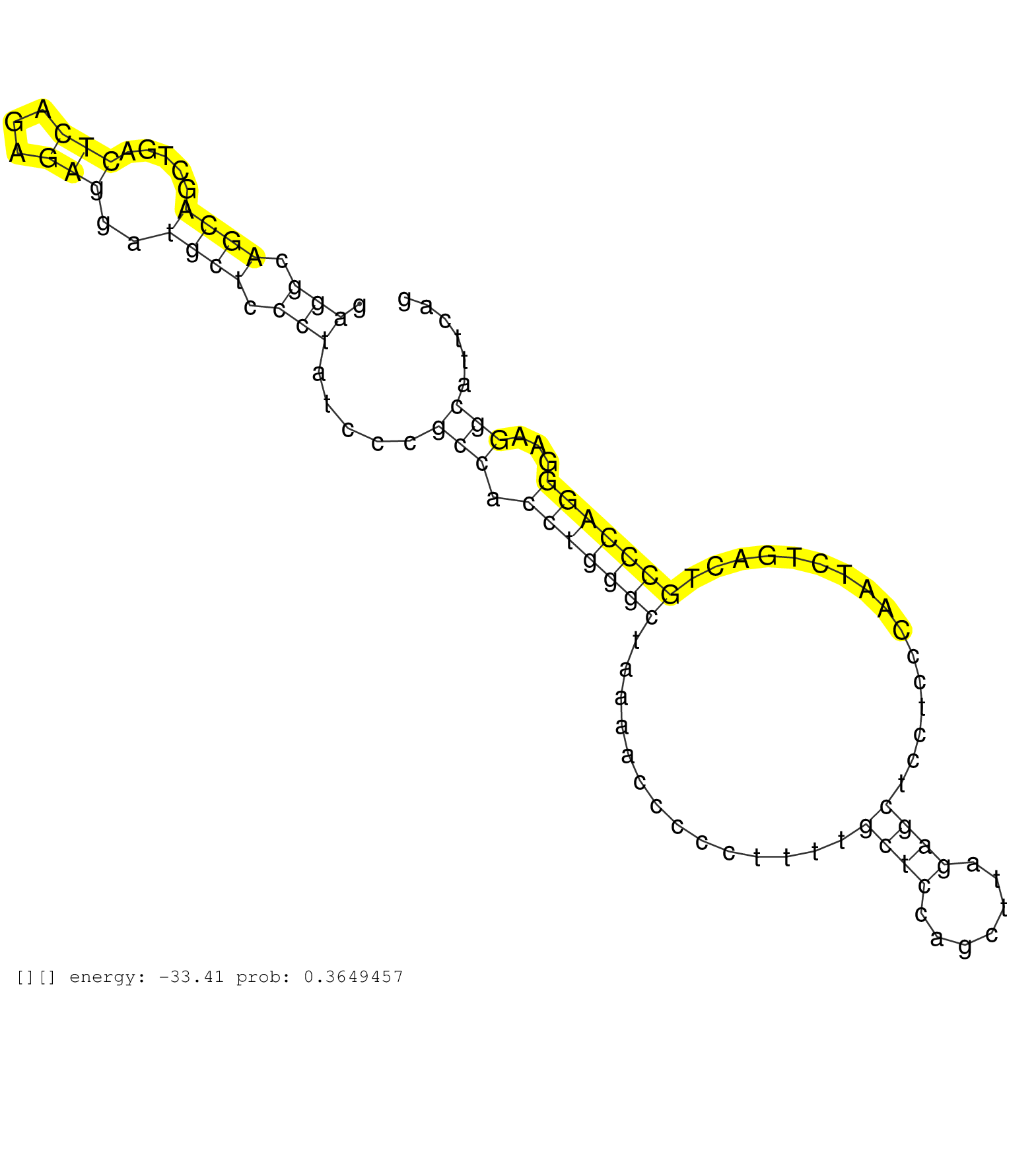
| GCTCCAAGGTTACAGCAACCGAACAACCCACCTCCTCACCATCTGAGGAGACAGTCGGCACCACTCTGTCAGGGAGACAGACCAAGCCACAGCCCCTCAATTAGAGGGGCGTGGGCTTCTTGCCATACAACTCCAGTTTACATCTGGATGAAACCCCAAGGGACCAAATCTGTAGCTGACCACTCCAGAGGCAGCAGCTGACTCAGAGAGGATGCTCCCTATCCCGCCACCTGGGCTAAAACCCCCTTTTGCTCCAGCTTAGAGCTCCTCCCAATCTGACTGCCCAGGGAAGGCATTCAGCCATGCCTTATCCATATTTAACTTTGGAACCTAAAAATGCCCCATTGGGT ............................................................................................................................................................................................(((.((((.....(((...)))..)))).))).....(((.(((((((..............((((.......))))................)))))))...)))........................................................ ...........................................................................................................................................................................................188.............................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................AGCAGCTGACTCAGAAC............................................................................................................................................. | 17 | 2.00 | 0.00 | 2.00 | - | - | |
| ................................................................................................................................................................................................AGCAGCTGACTCAGAACT............................................................................................................................................ | 18 | 2.00 | 0.00 | 1.00 | 1.00 | - | |
| ...............................................................................................................................................................................................................................................................................CAATCTGACTGCCCAGGGAAG.......................................................... | 21 | 1 | 2.00 | 2.00 | 2.00 | - | - |
| ...........................................TGAGGAGACAGTCGGCACCACTCT........................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .....................................................................................................................................................................AAATCTGTAGCTGACC......................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..........................................................................................................................................................................................................................................................................................................................TATTTAACTTTGGAACCTAAAAATGCCCCATT.... | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .............................................................................................................................................................................AGCTGACCACTCCAGAGGCAGCAGCTGACTCAGA............................................................................................................................................... | 34 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...................................................................................................................................................................................................................................................................TAGAGCTCCTCCCAATCTGACTGCCCAGGGAA........................................................... | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...............................................................................................................................................................................................................................................................AGCTTAGAGCTCCTCCCAATCTGACTGC................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .................................................GACAGTCGGCACCACTCTGTCAGGGAGAC................................................................................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ............................................GAGGAGACAGTCGGCACCACTCTGTCAGGGAGA................................................................................................................................................................................................................................................................................. | 33 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ................................................AGACAGTCGGCACCACTCTGTCAGGGAGACA............................................................................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .................................................GACAGTCGGCACCACTCTGTCAGGGAGACA............................................................................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ............................................GAGGAGACAGTCGGCACCACTCTGTC........................................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...........................................................................................................................................................................................................................................................................................................................ATTTAACTTTGGAACC................... | 16 | 2 | 0.50 | 0.50 | - | - | 0.50 |
| GCTCCAAGGTTACAGCAACCGAACAACCCACCTCCTCACCATCTGAGGAGACAGTCGGCACCACTCTGTCAGGGAGACAGACCAAGCCACAGCCCCTCAATTAGAGGGGCGTGGGCTTCTTGCCATACAACTCCAGTTTACATCTGGATGAAACCCCAAGGGACCAAATCTGTAGCTGACCACTCCAGAGGCAGCAGCTGACTCAGAGAGGATGCTCCCTATCCCGCCACCTGGGCTAAAACCCCCTTTTGCTCCAGCTTAGAGCTCCTCCCAATCTGACTGCCCAGGGAAGGCATTCAGCCATGCCTTATCCATATTTAACTTTGGAACCTAAAAATGCCCCATTGGGT ............................................................................................................................................................................................(((.((((.....(((...)))..)))).))).....(((.(((((((..............((((.......))))................)))))))...)))........................................................ ...........................................................................................................................................................................................188.............................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|