| (1) HEART | (1) KIDNEY | (2) OTHER |
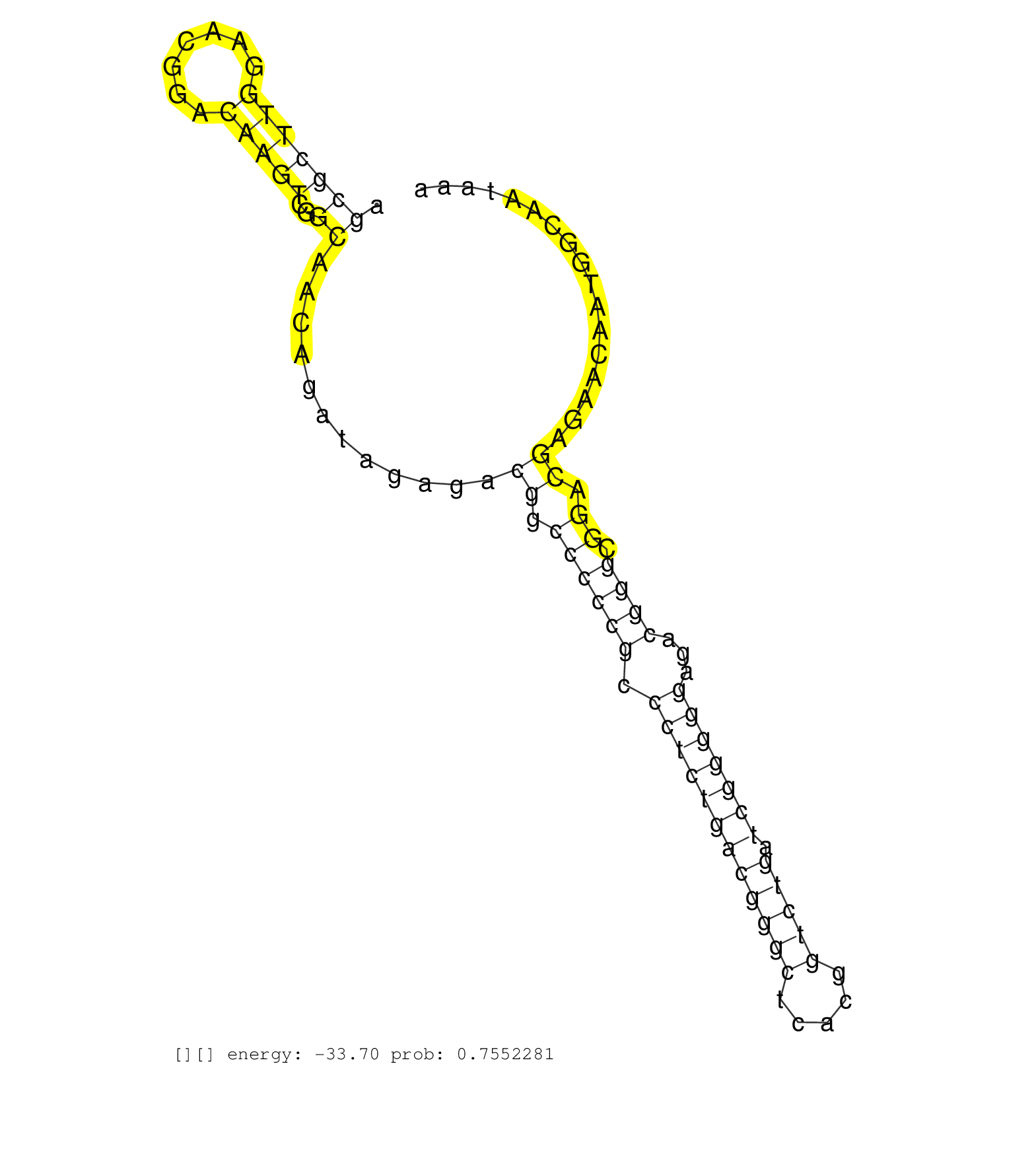
| CGGGAGAACCGCGACTACGTCCCTCAGTGCTTAGAGAACGGCCAGTTCCGGTAAGATTCATTCGTTCGATGGTATCTGTCGAGCGCCTACTATGTGCAGAGCACTGGACTAAGCGCTTGGAACGGACAAGTCGGCAACAGATAGAGACGGCCCCCGCCCTCTGACGGGCTCACGGTCTGATCGGGGGAGACGGGCGGACGAGAACAATGGCAATAAATAGAGTCGAGGGGAAGAGCATCTCGTAAAAACAGTGGCGAATAAATAGAATCGGGGTAATGTACGTCTCGATAAACCAAATGAATAGGGCGGTGGAGATCTATACAGTTGATATTCAGATGTCGGACCCTCTG ................................................................................................................(((((((.......)))))..))............((.((((((.((((((((((((.....))))).)))))))...)))).)).))...................................................................................................................................................... ...............................................................................................................112......................................................................................................217................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................TTGGAACGGACAAGTCGGCCACG................................................................................................................................................................................................................... | 23 | 3.00 | 0.00 | 3.00 | - | - | - | |
| ....................................................................................................................TTGGAACGGACAAGTCGGCACCG................................................................................................................................................................................................................... | 23 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .........................................................................................................GGACTAAGCGCTTGGAACGGACAAGCC.......................................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | - | - | - | 1.00 | |
| ..................................................................................................................................................................................................CGGACGAGAACAATGGAAA......................................................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ..............................................................................................................................CAAGTCGGCAACAGATAGAGACGGATAA.................................................................................................................................................................................................... | 28 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ..................................................................................................................GCTTGGAACGGACAAGTA.......................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ....................................................................................................................TTGGAACGGACAAGTCGGCAACAGTAAA.............................................................................................................................................................................................................. | 28 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ....................................................................................................................TTGGAACGGACAAGTCGGCAAAAGA................................................................................................................................................................................................................. | 25 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .....................................................................................................................................................................................................ACGAGAACAATGGCAATAAATAGAGAAA............................................................................................................................. | 28 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ...................................................................................................AGCACTGGACTAAGCGCTTGGAACGGT................................................................................................................................................................................................................................ | 27 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ....................................................................................................................TTGGAACGGACAAGTCGGA....................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ....................................................................................................................TTGGAACGGACAAGTCGGCAACAGAGACA............................................................................................................................................................................................................. | 29 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ...................................................................................................................................................................ACGGGCTCACGGTCTGATCGGGGGATACA.............................................................................................................................................................. | 29 | 1.00 | 0.00 | - | - | 1.00 | - | |
| .....................................................................................................................................................................................................ACGAGAACAATGGCAATAACCAG.................................................................................................................................. | 23 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ................................................................................................CAGAGCACTGGACTAAGCGCTATA...................................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ....................................................................................................................TTGGAACGGACAAGTCGGCAT..................................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ...............................................................................................................................................................................................................................................................GAATAAATAGAATCGGGG............................................................................. | 18 | 12 | 0.08 | 0.08 | - | 0.08 | - | - |
| CGGGAGAACCGCGACTACGTCCCTCAGTGCTTAGAGAACGGCCAGTTCCGGTAAGATTCATTCGTTCGATGGTATCTGTCGAGCGCCTACTATGTGCAGAGCACTGGACTAAGCGCTTGGAACGGACAAGTCGGCAACAGATAGAGACGGCCCCCGCCCTCTGACGGGCTCACGGTCTGATCGGGGGAGACGGGCGGACGAGAACAATGGCAATAAATAGAGTCGAGGGGAAGAGCATCTCGTAAAAACAGTGGCGAATAAATAGAATCGGGGTAATGTACGTCTCGATAAACCAAATGAATAGGGCGGTGGAGATCTATACAGTTGATATTCAGATGTCGGACCCTCTG ................................................................................................................(((((((.......)))))..))............((.((((((.((((((((((((.....))))).)))))))...)))).)).))...................................................................................................................................................... ...............................................................................................................112......................................................................................................217................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................aaaCTATCTGTTGCCGACTTGTAAA........................................................................................................................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ................................................................................................................................................................................................aTATTGCCATTGTTCTCGTCCGCA...................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ................gAGCACTGAGGGACGG.............................................................................................................................................................................................................................................................................................................................. | 16 | 1.00 | 0.00 | - | - | 1.00 | - |