| (1) BRAIN | (1) HEART | (1) KIDNEY | (2) OTHER |
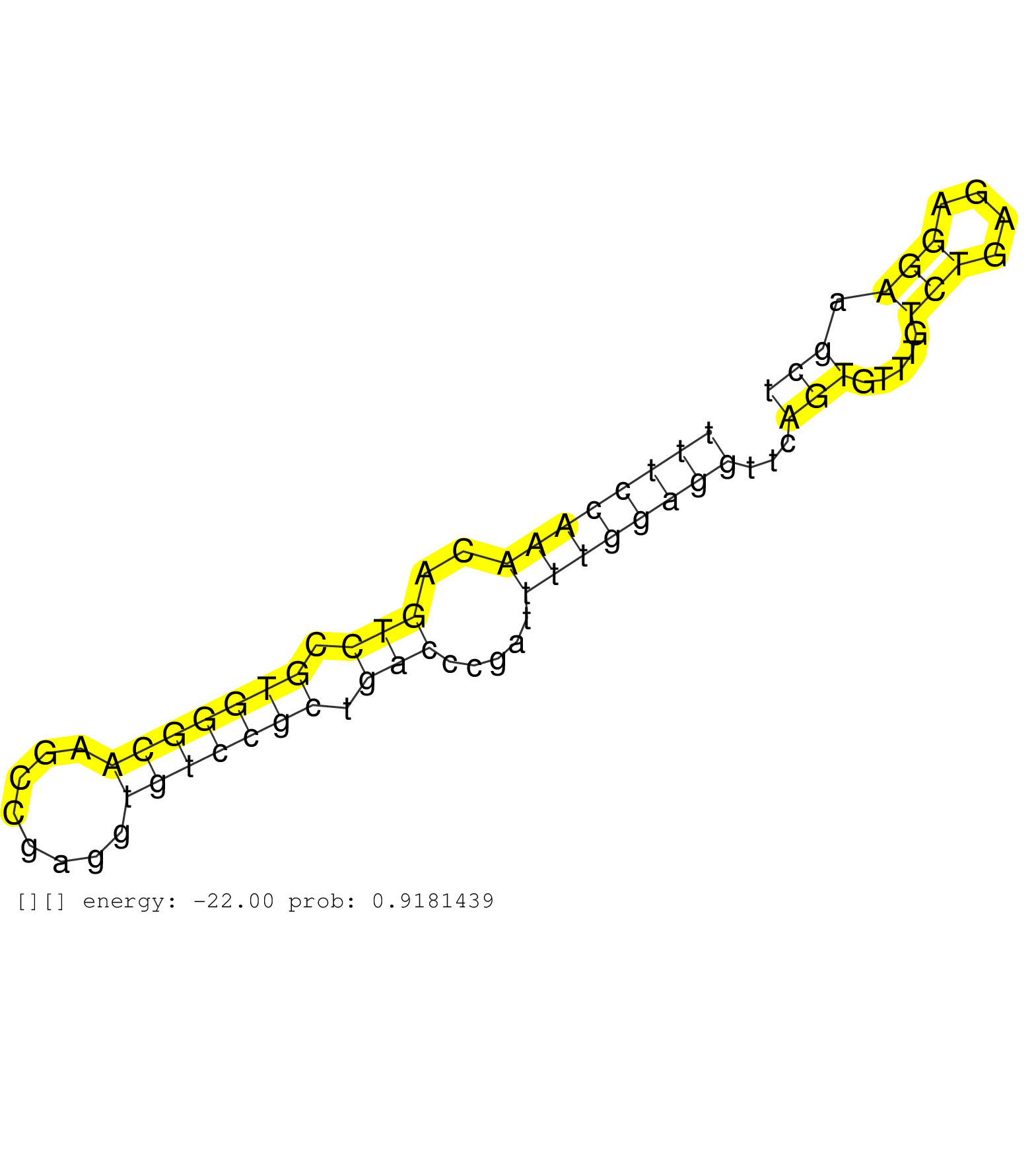
| CAGAACGAGGACCTGGGACTCAACGACATGAAGACAGAGGGCTACGAGGCTGGCGGGCCCCCCCGGCGGTAGCCCCCGGCCCGCCGGAGGGGCAGGGGCGACGATATAATTATTACTGTTGTAATAGGTCCAAAGGGAGCAGCCCGGGGATCGAGGCCGGGCTGCCCGGAGGGGGCTCGACTGGAGCCAGGAAGCCGCCCGCCCTTTCCAAACAGTCCGTGGGCAAGCCGAGGTGTCCGCTGACCCGATTTTGGAGGTTCAGTGTTTGTCTGAGAGGAAGCTCGCCGTCGGACAGGTTTAAAAGCGTTTTTCCCTCAAGCCACC ............................................................................................................................................................................................................((((((((..(((.(((((((........))))))).))).....))))))))...(((.....(((....))).))).......................................... ............................................................................................................................................................................................................205..........................................................................282........................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553592(SRX182798) source: Brain. (Brain) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................TACTGTTGTAATAGGTCCAAAGGGAGCAGC..................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ......GAGGACCTGGGACTCAACGAC......................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| .................................................................................................................................................................................................................AAACAGTCCGTGGGCAAGCC............................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ......GAGGACCTGGGACTCAACGACATGAAGACAGA.............................................................................................................................................................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .......AGGACCTGGGACTCAACGACAT....................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................................................................................................AGTGTTTGTCTGAGAGGA.............................................. | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..............GGGACTCAACGACATGAAGACAGAGGGCT......................................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .............TGGGACTCAACGACATGAAGAC................................................................................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................................................CAGTCCGTGGGCAAGCCGAGGT.......................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..GAACGAGGACCTGGGACTC............................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..........................................................................................................................AATAGGTCCAAAGGGA.......................................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..............GGGACTCAACGACATGAAGACA................................................................................................................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................................................................................................................CCGTCGGACAGGTTTAAAAGCGTTTTT............. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - |
| ...AACGAGGACCTGGGACTCAACGAC......................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| .............................................................................................................................AGGTCCAAAGGGAGCAGC..................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................AGCTCGCCGTCGGAC............................... | 15 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - |
| .............................................................................................................................................................CGGGCTGCCCGGAGGG....................................................................................................................................................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 |
| CAGAACGAGGACCTGGGACTCAACGACATGAAGACAGAGGGCTACGAGGCTGGCGGGCCCCCCCGGCGGTAGCCCCCGGCCCGCCGGAGGGGCAGGGGCGACGATATAATTATTACTGTTGTAATAGGTCCAAAGGGAGCAGCCCGGGGATCGAGGCCGGGCTGCCCGGAGGGGGCTCGACTGGAGCCAGGAAGCCGCCCGCCCTTTCCAAACAGTCCGTGGGCAAGCCGAGGTGTCCGCTGACCCGATTTTGGAGGTTCAGTGTTTGTCTGAGAGGAAGCTCGCCGTCGGACAGGTTTAAAAGCGTTTTTCCCTCAAGCCACC ............................................................................................................................................................................................................((((((((..(((.(((((((........))))))).))).....))))))))...(((.....(((....))).))).......................................... ............................................................................................................................................................................................................205..........................................................................282........................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553592(SRX182798) source: Brain. (Brain) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|