| (1) KIDNEY | (2) OTHER |
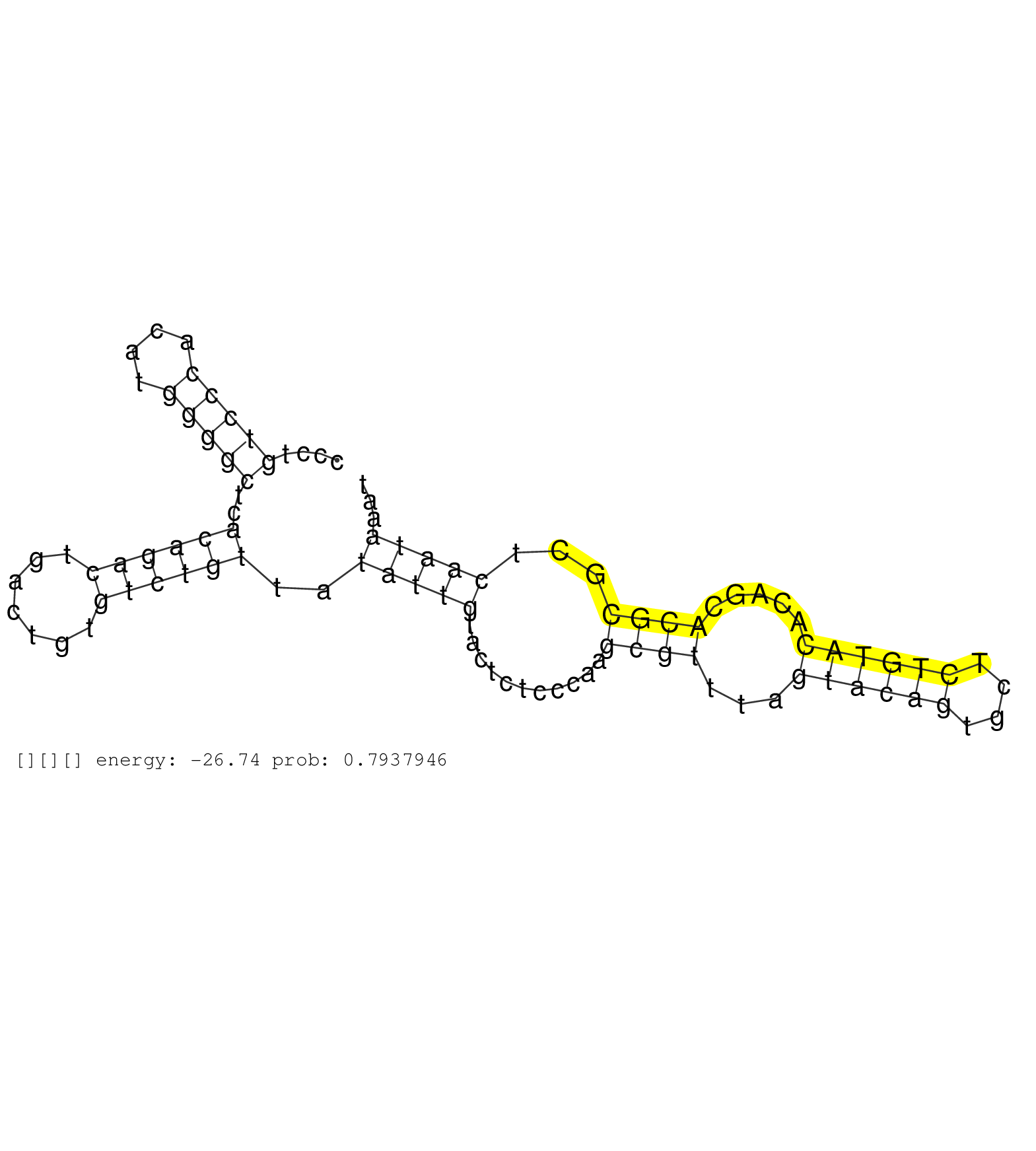
| AGCCCTCCGTCTTCACTCGCGTCTCCAACTACAACAGCTGGATCAATGAGGTGAGGCCCCTTGGATCCTGCACTGCTCCTCAAGACCTCTAGACTGGAAGCTCCTTGTGGGCAGGGAATGTTTTCTTTCTTTGTGGTATTTAAGCGCTTCTTATGTGCTAAACCCCGGGGTAGACACGTGACAACCAGGCCGGATCTGGTCCCTGTCCCACATGGGGCTCACAGACTGACTGTGTCTGTTATATTGTACTCTCCCAAGCGTTTAGTACAGTGCTCTGTACACAGCACGCGCTCAATAAATAAGTGATGGACTGACTGAAGGGTGGACAGAGAGGTTGACTCTGACAGGCT ............................................................................................................................................................................................................(((((....)))))..((((((.......))))))..(((((...........((((...((((((....)))))).....))))...)))))..................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|
| .........................................................................................TAGACTGGAAGCTCCTTGTGGGCAAAA.......................................................................................................................................................................................................................................... | 27 | 3.00 | 0.00 | 3.00 | - | - | |
| .........................................................................................TAGACTGGAAGCTCCTTGTGGGCAATA.......................................................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | 1.00 | - | - | |
| .........................................................................................TAGACTGGAAGCTCCTTGTGAAA.............................................................................................................................................................................................................................................. | 23 | 1.00 | 0.00 | 1.00 | - | - | |
| .........................................................................................TAGACTGGAAGCTCCTTGTGGGCAGTT.......................................................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | 1.00 | - | - | |
| .........................................................................................TAGACTGGAAGCTCCTTGTGGCAT............................................................................................................................................................................................................................................. | 24 | 1.00 | 0.00 | 1.00 | - | - | |
| .........................................................................................TAGACTGGAAGCTCCTTGTGCGC.............................................................................................................................................................................................................................................. | 23 | 1.00 | 0.00 | 1.00 | - | - | |
| .........................................................................................TAGACTGGAAGCTCCTTGTGGGCAGCT.......................................................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | 1.00 | - | - | |
| .................................................................................................................................................................................................................................................................................TCTGTACACAGCACGCAC........................................................... | 18 | 1.00 | 0.00 | 1.00 | - | - | |
| .......................................................................................TCTAGACTGGAAGCTCCTTGTGGGCATTT.......................................................................................................................................................................................................................................... | 29 | 1.00 | 0.00 | 1.00 | - | - | |
| .........................................................................................TAGACTGGAAGCTCCTTGTGGGCAGTTTT........................................................................................................................................................................................................................................ | 29 | 1.00 | 0.00 | 1.00 | - | - | |
| .........................................................................................TAGACTGGAAGCTCCTTGTGGCAGT............................................................................................................................................................................................................................................ | 25 | 1.00 | 0.00 | 1.00 | - | - | |
| .........................................................................................TAGACTGGAAGCTCCTTGTGGGCACA........................................................................................................................................................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | - | |
| ......................CTCCAACTACAACAGC........................................................................................................................................................................................................................................................................................................................ | 16 | 3 | 0.33 | 0.33 | - | 0.33 | - |
| AGCCCTCCGTCTTCACTCGCGTCTCCAACTACAACAGCTGGATCAATGAGGTGAGGCCCCTTGGATCCTGCACTGCTCCTCAAGACCTCTAGACTGGAAGCTCCTTGTGGGCAGGGAATGTTTTCTTTCTTTGTGGTATTTAAGCGCTTCTTATGTGCTAAACCCCGGGGTAGACACGTGACAACCAGGCCGGATCTGGTCCCTGTCCCACATGGGGCTCACAGACTGACTGTGTCTGTTATATTGTACTCTCCCAAGCGTTTAGTACAGTGCTCTGTACACAGCACGCGCTCAATAAATAAGTGATGGACTGACTGAAGGGTGGACAGAGAGGTTGACTCTGACAGGCT ............................................................................................................................................................................................................(((((....)))))..((((((.......))))))..(((((...........((((...((((((....)))))).....))))...)))))..................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|
| ............................................................................aggaTAGAGGTCTTGAGGA............................................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | |
| ..........................................................GTGCAGGATCCAAGG..................................................................................................................................................................................................................................................................................... | 15 | 6 | 0.17 | 0.17 | - | - | 0.17 |