| (1) HEART | (1) KIDNEY | (1) OTHER |
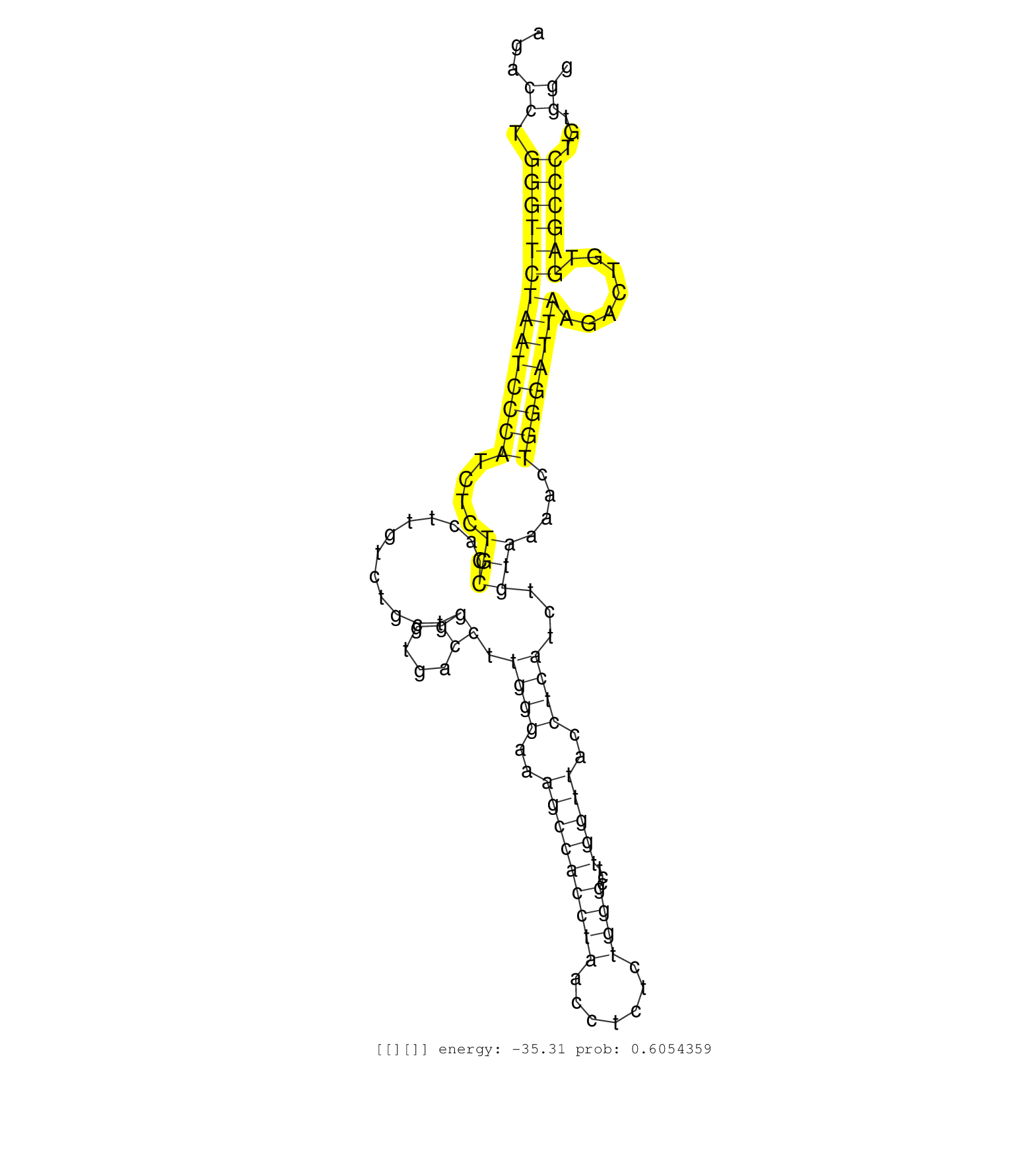
| GTGTAGTAATCCTATCAGAGATCATGATTGCCTGTTTGGAGCACAATGAGGTGATTATTTGTTTTATGTGAAATAAGTCTTTTCATTGGGAAATGCAGGGGTCTGATAGTCTGATAAAAGCAGCGTGGCCTACTGTAGAGAGCCTGGACCTGGGATTCAAAAAGACCTGGGTTCTAATCCCATCTCTGCCACTTGTCTGCTGGGTGACCTTGGGAAAGCCACCTAACCTCTCTGGGCCTTGGTTACCTCATCTGTAAAACTGGGATTAAGACTGTGAGCCCTGTGGGGGTCAAGTTAACCTGATCATAGTAAGCACTTAAATATTATTAAAAAAATAGTCTGGGAAGCAG .....................................................................................................................................................................((.((((((((((((((....(((............((....)).((((..(((((((((.......))))...)))))..))))...)))....)))))))).......))))))...))................................................................ ..................................................................................................................................................................163.........................................................................................................................287............................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................................................CATCTGTAAAACTGGGATTAAGACGG............................................................................ | 26 | 2.00 | 0.00 | 2.00 | - | - | |
| ...............CAGAGATCATGATTGCCTGTTTGG....................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ....................................................................................................................................................................................................................................................................TGGGATTAAGACTGTGAGCGGCA................................................................... | 23 | 1.00 | 0.00 | 1.00 | - | - | |
| ....................................................................................................................................................................................................................................................................TGGGATTAAGACTGTGCAA....................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | |
| ....................ATCATGATTGCCTGTTTG........................................................................................................................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .......................................................................................................................................................................................................................................................TCATCTGTAAAACTGGGATTAAGACGGTA.......................................................................... | 29 | 1.00 | 0.00 | 1.00 | - | - | |
| .........................GATTGCCTGTTTGGAGC.................................................................................................................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .......................................................................................................................................................................................................................................................TCATCTGTAAAACTGGGATTAAGACGGT........................................................................... | 28 | 1.00 | 0.00 | 1.00 | - | - | |
| ....................................................................................................................................................................................................................................................................TGGGATTAAGACTGTGAGAAAA.................................................................... | 22 | 1.00 | 0.00 | 1.00 | - | - | |
| .......................................................................................................................................................................................................................................................TCATCTGTAAAACTGGGATTAAGAAA............................................................................. | 26 | 1.00 | 0.00 | 1.00 | - | - | |
| .......................................................................................................................................................................................................................................................TCATCTGTAAAACTGGGATTAAGACGGGG.......................................................................... | 29 | 1.00 | 0.00 | 1.00 | - | - | |
| ................AGAGATCATGATTGCCTGTTTGGAGCACAAT............................................................................................................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ....................................................................................................................................................................................................................................................................TGGGATTAAGACTGTGAGAACA.................................................................... | 22 | 1.00 | 0.00 | 1.00 | - | - | |
| .......................................................................................................................................................................TGGGTTCTAATCCCATCTCGGCA................................................................................................................................................................ | 23 | 1.00 | 0.00 | 1.00 | - | - | |
| .......................ATGATTGCCTGTTTG........................................................................................................................................................................................................................................................................................................................ | 15 | 2 | 0.50 | 0.50 | - | 0.50 | - |
| GTGTAGTAATCCTATCAGAGATCATGATTGCCTGTTTGGAGCACAATGAGGTGATTATTTGTTTTATGTGAAATAAGTCTTTTCATTGGGAAATGCAGGGGTCTGATAGTCTGATAAAAGCAGCGTGGCCTACTGTAGAGAGCCTGGACCTGGGATTCAAAAAGACCTGGGTTCTAATCCCATCTCTGCCACTTGTCTGCTGGGTGACCTTGGGAAAGCCACCTAACCTCTCTGGGCCTTGGTTACCTCATCTGTAAAACTGGGATTAAGACTGTGAGCCCTGTGGGGGTCAAGTTAACCTGATCATAGTAAGCACTTAAATATTATTAAAAAAATAGTCTGGGAAGCAG .....................................................................................................................................................................((.((((((((((((((....(((............((....)).((((..(((((((((.......))))...)))))..))))...)))....)))))))).......))))))...))................................................................ ..................................................................................................................................................................163.........................................................................................................................287............................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|