| (1) HEART | (1) KIDNEY | (2) OTHER |
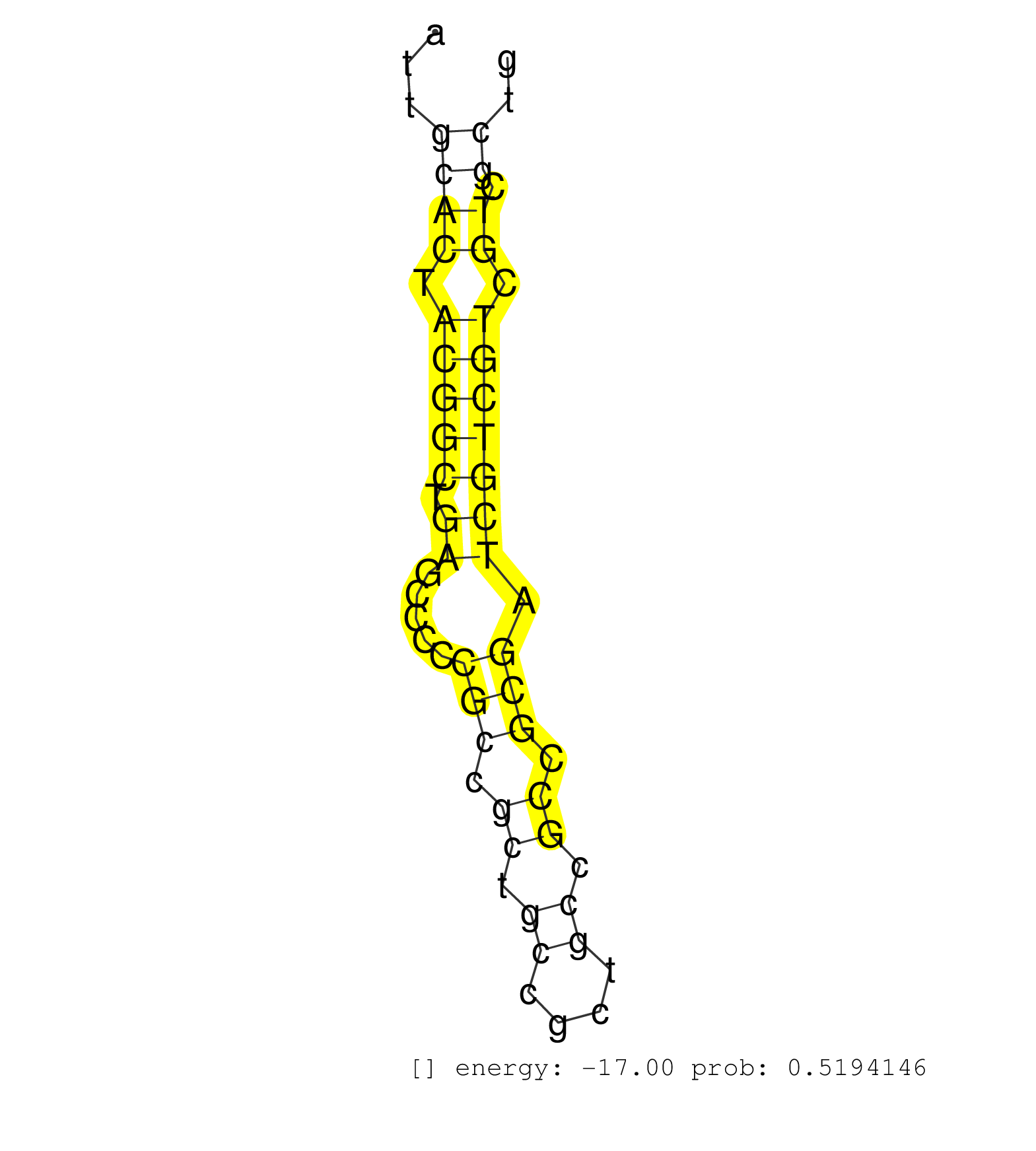
| AAAGCGTTGCTTTGAAAACACTTTCCAAGAACTCATCGCGGCCGCCAACGGCCAGTTGTAGTATTGGCAGGAAACATGCAGGACATGGCCCAGAAACTGACCCGAGAGCAAATTGCACTACGGCTGAGCCCCCGCCGCTGCCGCTGCCGCCGCGATCGTCGTCGTCGCTGCCGTGACCCTCCCGTTTGGCATCGGGGCCCCAGCCCGGAGGCGGGCGAGGCTGCTGCAGGTACACTGTTACTTGAAGGGGAGAACGTTTTACTCACCGAGAGCTACGGCTGCCGTCGGAGGCTTTGATCTGTGAAGAATTTTATTTTAGATCGAGGAACACCATTGGGTGTGTGATGTTC ..................................................................................................................((((.(((((.((.....(((.((.((....)).)).))).))))))).)).))...................................................................................................................................................................................... ...............................................................................................................112.......................................................170.................................................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................ACACTGTTACTTGAAGGGGAGAACGT............................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ................................................CGGCCAGTTGTAGTATTGGCAGGAAACATGC............................................................................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ......................................................................................................................................................CGCGATCGTCGTCGTCGCTGCCGTGACC............................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| .............................................................................................................................................................................................................................................................................................................TGAAGAATTTTATTTTAGATCGAGGAACACC.................. | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ....................................................................................................................ACTACGGCTGAGCCCCCG........................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .......................................................................................................................................................................................................................................................................................................................TATTTTAGATCGAGGAACACCATTGGGT........... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................TTTTAGATCGAGGAACACCATTGGGTGTGTGT..... | 32 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ...............................................ACGGCCAGTTGTAGTATTG............................................................................................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ................................................................................................................................................................................................................................................................................................................AGAATTTTATTTTAGATCGAGGAACACC.................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ....................................................................................................................................................................................................................................................................................................................TTTTATTTTAGATCGAGGAACACC.................. | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ....................................................................................................................................................GCCGCGATCGTCGTCGTC........................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .................................................................................................................................................GCCGCCGCGATCGTCG............................................................................................................................................................................................. | 16 | 2 | 0.50 | 0.50 | - | - | - | 0.50 |
| ........................................................................................................................................................................................................................................CACTGTTACTTGAAGGG..................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | 0.50 | - |
| .....................................................................................................................................................................................................CCCCAGCCCGGAGGC.......................................................................................................................................... | 15 | 6 | 0.17 | 0.17 | - | 0.17 | - | - |
| AAAGCGTTGCTTTGAAAACACTTTCCAAGAACTCATCGCGGCCGCCAACGGCCAGTTGTAGTATTGGCAGGAAACATGCAGGACATGGCCCAGAAACTGACCCGAGAGCAAATTGCACTACGGCTGAGCCCCCGCCGCTGCCGCTGCCGCCGCGATCGTCGTCGTCGCTGCCGTGACCCTCCCGTTTGGCATCGGGGCCCCAGCCCGGAGGCGGGCGAGGCTGCTGCAGGTACACTGTTACTTGAAGGGGAGAACGTTTTACTCACCGAGAGCTACGGCTGCCGTCGGAGGCTTTGATCTGTGAAGAATTTTATTTTAGATCGAGGAACACCATTGGGTGTGTGATGTTC ..................................................................................................................((((.(((((.((.....(((.((.((....)).)).))).))))))).)).))...................................................................................................................................................................................... ...............................................................................................................112.......................................................170.................................................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|