| (1) BRAIN | (1) HEART | (1) KIDNEY | (2) OTHER |
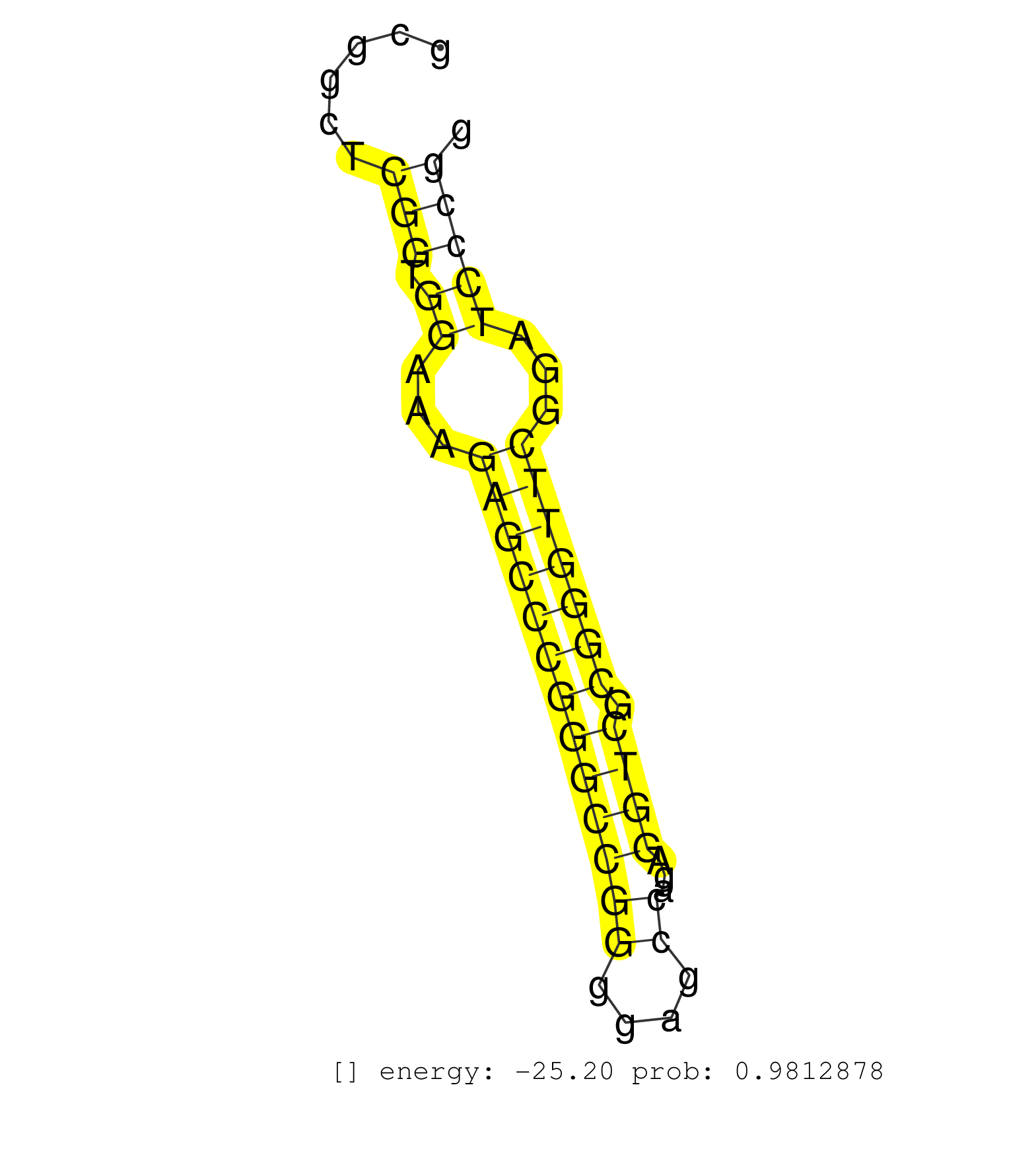
| TGTTGGTCTTGGCCATGGACTGCGGCGTCAACTGCTTCATGCTCGACCCGGTGAGGAGGACGCCCCCGCCCCCCGTCCGAGGCCGGGGAGCGGGGAGGCGGGGGGCCTGGGCCTCGCAAGGGGCGTAGTGGCGGTATTCATTAGAGAAGCAGCGCGGCTCGGTGGAAAGAGCCCGGGCCGGGGAGCCAGAGGTCGCGGGTTCGGATCCCGGCTCGGCCGCCTGTCAGCCGGGTGACCGTGGGCGGGTCGCTTCACTTCTCTGGGCCTCAGTTCCCTCGACCAAAATGGGGATGAAGGCGGGGAGCCTCACGAGACAGCCCGATGAGCCCGGATCTACCCCGGCGCTTAGA ...............................................................................................................................................................(((.((...(((((((((((((....))...)))).)))))))...)))))............................................................................................................................................ .........................................................................................................................................................154......................................................211......................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553592(SRX182798) source: Brain. (Brain) |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................AGGTCGCGGGTTCGGTCC............................................................................................................................................... | 18 | 11.00 | 0.00 | 6.00 | 2.00 | 1.00 | 1.00 | 1.00 | |
| .............................................................................................................................................................................................AGGTCGCGGGTTCGGTCCC.............................................................................................................................................. | 19 | 3.00 | 0.00 | 3.00 | - | - | - | - | |
| ...............TGGACTGCGGCGTCAAC.............................................................................................................................................................................................................................................................................................................................. | 17 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - |
| .............................................................................................................................................................................................AGGTCGCGGGTTCGGT................................................................................................................................................. | 16 | 2.00 | 0.00 | 1.00 | - | - | 1.00 | - | |
| .........TGGCCATGGACTGCGGCGTCAAC.............................................................................................................................................................................................................................................................................................................................. | 23 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - |
| .............CATGGACTGCGGCGTCAACTGC........................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..............ATGGACTGCGGCGTCAACTGCT.......................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..............................................................................................................................................................TCGGTGGAAAGAGCCCGGGCCAG......................................................................................................................................................................... | 23 | 1.00 | 0.00 | - | 1.00 | - | - | - | |
| ..TTGGTCTTGGCCATGGACTGCGGCGTCAAC.............................................................................................................................................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ......................CGGCGTCAACTGCTTCATGCTCGACCCGGC.......................................................................................................................................................................................................................................................................................................... | 30 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| ..............ATGGACTGCGGCGTCAACT............................................................................................................................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .GTTGGTCTTGGCCATGGACTGCGGCGT.................................................................................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..............ATGGACTGCGGCGTC................................................................................................................................................................................................................................................................................................................................. | 15 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..............ATGGACTGCGGCGTCAACTGC........................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| TGTTGGTCTTGGCCATGGACTGCGGCGTCAACTGCTTCATGCTCGACCCGGTGAGGAGGACGCCCCCGCCCCCCGTCCGAGGCCGGGGAGCGGGGAGGCGGGGGGCCTGGGCCTCGCAAGGGGCGTAGTGGCGGTATTCATTAGAGAAGCAGCGCGGCTCGGTGGAAAGAGCCCGGGCCGGGGAGCCAGAGGTCGCGGGTTCGGATCCCGGCTCGGCCGCCTGTCAGCCGGGTGACCGTGGGCGGGTCGCTTCACTTCTCTGGGCCTCAGTTCCCTCGACCAAAATGGGGATGAAGGCGGGGAGCCTCACGAGACAGCCCGATGAGCCCGGATCTACCCCGGCGCTTAGA ...............................................................................................................................................................(((.((...(((((((((((((....))...)))).)))))))...)))))............................................................................................................................................ .........................................................................................................................................................154......................................................211......................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553592(SRX182798) source: Brain. (Brain) |
|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................TCCGAACCCGCGACCTCT................................................................................................................................................. | 18 | 10 | 0.10 | 0.10 | - | 0.10 | - | - | - |