| (1) HEART | (1) KIDNEY | (1) OTHER |
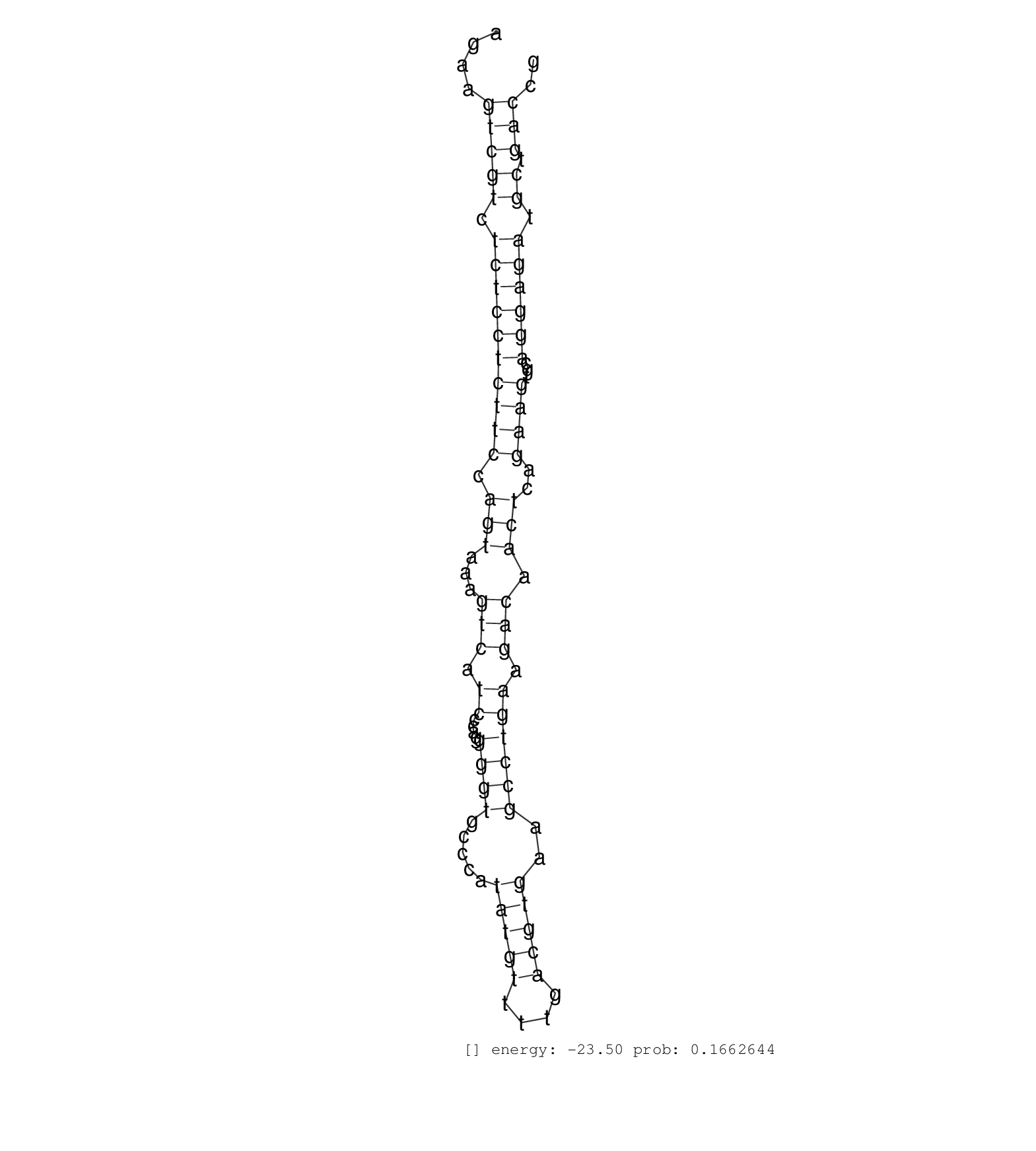
| CGTTAGTCATACCAGCGTCTGCAGGAGCATCAGCTGCAAGCTGGTCAAAAGAGAGAAAAGATTAGAAGAGAACGAGATGATAAACTTATCGCTTCATCCCTCCAAGAAGAGTGGGCCTCAGAAGCAGCTCCCCAGCAGCAACAGATGGAATTATTCTTTCATCAGAAAAAAACGAAGGCGGTACAGCAAGAAATCGGCCTGCGTGGTACCGGCCGCCTTGAGGCCTCCTCAGGAGCCTCATTCGGGAGCCCTACCGGAATCCTCTGGGACGGAAAGAAGTCGTCTCTCCTCTTCCAGTAAAGTCATCCCAGGGGTGCCCATATGTTTTGACGTGAAGCCTGAAGACAACTCAGAAGTGCAGGAGATGCTGACCGCATCCCGCTCCATGACCAGAATTAAGAAGCGGTCTCGGCGGGTCTTCTAT ......................................................................................................................................................................................................................................................................................(((((.((((((((((.(((...(((.((....((((.....(((((....)))))..)))))).))).)))..))))...)))))).)).))).................................................... ..................................................................................................................................................................................................................................................................................275................................................................................................374................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................................................................................................................................TATGTTTTGACGTGAAGCCTGAAGACAAC........................................................................... | 29 | 1 | 19.00 | 19.00 | 19.00 | - | - |
| ................................................................................................................................................................................................................................................................................................................................TATGTTTTGACGTGAAGCCTGAAGACAACT.......................................................................... | 30 | 1 | 4.00 | 4.00 | 4.00 | - | - |
| ..............................................................................................................................................................................................................................................................................................................................CATATGTTTTGACGTGAAGCCTGAAGA............................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - |
| .........................................TGGTCAAAAGAGAGAAAAGATTAGAAGAGA................................................................................................................................................................................................................................................................................................................................................................. | 30 | 1 | 2.00 | 2.00 | 2.00 | - | - |
| ................................................................................................................................................................................................................................................................................................................................TATGTTTTGACGTGAAGCCTGAAGACAA............................................................................ | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - |
| .........................................TGGTCAAAAGAGAGAAAAGATTAGAAGAGAAC............................................................................................................................................................................................................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .......................................................................................................................................................................................................................................................................................................................................TGACGTGAAGCCTGAAGACAACTCAGA...................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................TGAAGACAACTCAGAAGTGCAGGAGATGC........................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...........................................GTCAAAAGAGAGAAACTTC.......................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | 1.00 | |
| ...................................................................................................................................................................................................................................................................................................................................................TGAAGACAACTCAGAAGTGCAGGAGATG......................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ........................................................................................................AGAAGAGTGGGCCTCAGAAGC........................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .........................................................................................................................................................................................GCAAGAAATCGGCCTGCGTGGTACCGGC................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ....................................................................................................................................................................................................................................................................................................................................................GAAGACAACTCAGAAGTGCAGGAGATGCTG...................................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .....................................................................................................................................................................................................................................................................................................................................TTTGACGTGAAGCCTGAAGACAACTCAGAAG.................................................................... | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| CGTTAGTCATACCAGCGTCTGCAGGAGCATCAGCTGCAAGCTGGTCAAAAGAGAGAAAAGATTAGAAGAGAACGAGATGATAAACTTATCGCTTCATCCCTCCAAGAAGAGTGGGCCTCAGAAGCAGCTCCCCAGCAGCAACAGATGGAATTATTCTTTCATCAGAAAAAAACGAAGGCGGTACAGCAAGAAATCGGCCTGCGTGGTACCGGCCGCCTTGAGGCCTCCTCAGGAGCCTCATTCGGGAGCCCTACCGGAATCCTCTGGGACGGAAAGAAGTCGTCTCTCCTCTTCCAGTAAAGTCATCCCAGGGGTGCCCATATGTTTTGACGTGAAGCCTGAAGACAACTCAGAAGTGCAGGAGATGCTGACCGCATCCCGCTCCATGACCAGAATTAAGAAGCGGTCTCGGCGGGTCTTCTAT ......................................................................................................................................................................................................................................................................................(((((.((((((((((.(((...(((.((....((((.....(((((....)))))..)))))).))).)))..))))...)))))).)).))).................................................... ..................................................................................................................................................................................................................................................................................275................................................................................................374................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|