| (1) BRAIN | (1) HEART | (1) KIDNEY |
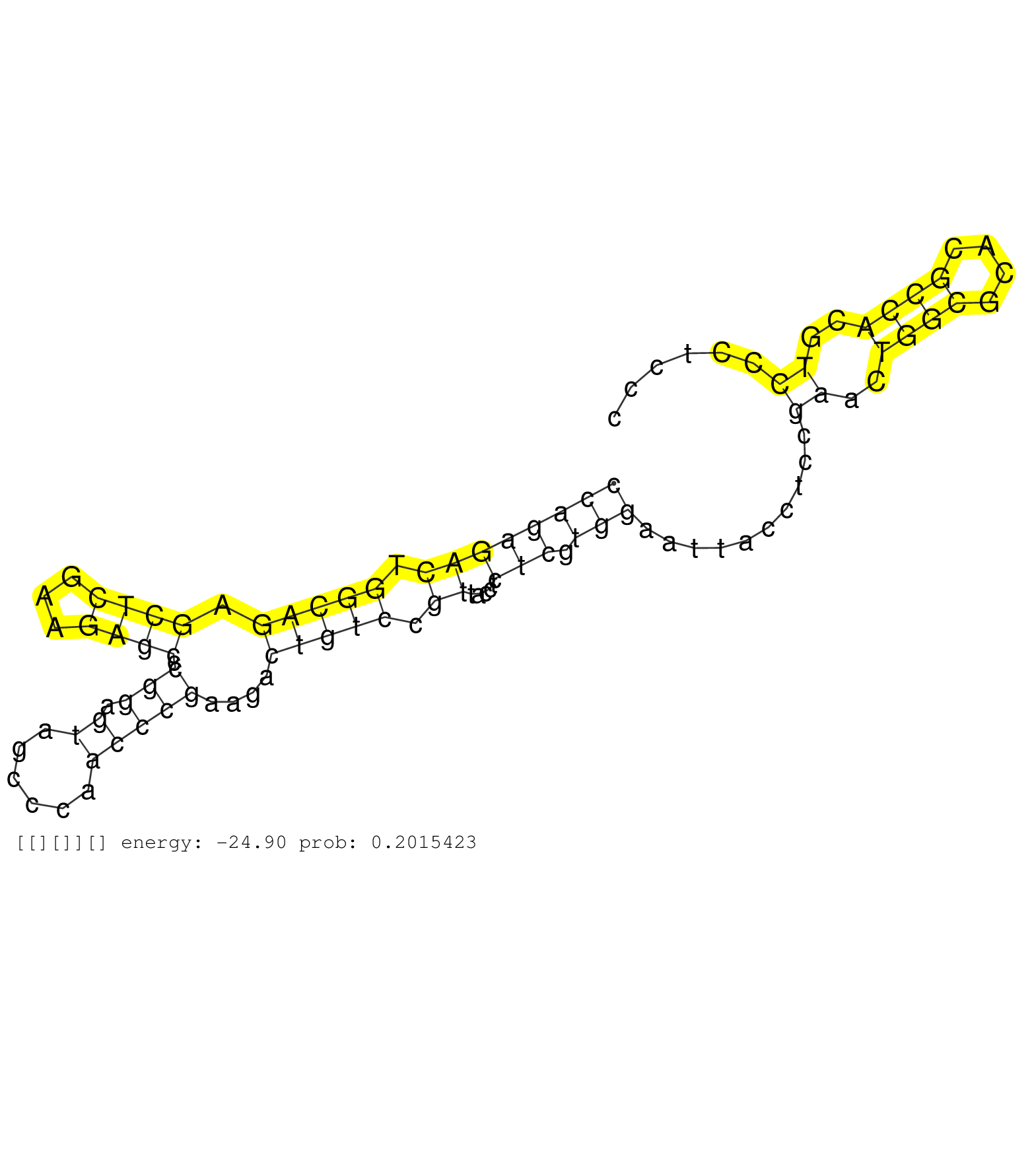
| CCGGCCCCTGCCTTCACCAGCCACCCCCCAGAGCCAAGAAGACGTCCAAGAGGTGGACCAGGGGAGGAACGTGGTCCCGGCGCTCCAGACCCAGGAGTCCCAGAGACTGGCAGAGCTCGAAGAGCCCGGAGTAGCCCAACCCGAAGACTGTCCGTTACCTCTCGTGGAATTACCTCCGAACTGGCGCACGCCACGTCCCTCCCTCTCCTTCCTAGCAGCGAGGGTGAACCGGGCCCCGAGGCCGCGGTTTCGCTCCCGGCCCCTCTGCCCCCATCGCCCCCTCCACCGCCCCCGCCGCCCCCTCCTCCACCCCCGCCTCTGCCCACGAAGGAGGACGGCGCGGAGCCGCT ...................................................................................................((((((((.(((((.((((...)))).(((.((......)))))....))))).)).....))).)))..........((..((((....))))..))......................................................................................................................................................... ...................................................................................................100.....................................................................................................203................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553592(SRX182798) source: Brain. (Brain) |
|---|---|---|---|---|---|---|---|
| ........................................................................................................GACTGGCAGAGCTCGAAGA................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .......................................................................................................AGACTGGCAGAGCTCGAAGAGCCC............................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...............................................................................................................................................AAGACTGTCCGTTAC................................................................................................................................................................................................ | 15 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...............................................................................................AGTCCCAGAGACTGGAAC............................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | 1.00 | |
| ....................................................................................................CAGAGACTGGCAGAGCTCGAAGA................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...........................................................................................CAGGAGTCCCAGAGACTGGCAG............................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ....................................................................................................CAGAGACTGGCAGAGCTC........................................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ............................................................................................AGGAGTCCCAGAGACTGGC............................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...............................................................................................AGTCCCAGAGACTGGCAGAGCTCGA...................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .....................................................................................................AGAGACTGGCAGAGCTCGAAGAGCCCGGA............................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .............................................................................................................................................CGAAGACTGTCCGTTACCTCTCGTGGACATT.................................................................................................................................................................................. | 31 | 1.00 | 0.00 | 1.00 | - | - | |
| ....................................................................................................................................................................................CTGGCGCACGCCACGTCCC....................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ......................................................................................................GAGACTGGCAGAGCT......................................................................................................................................................................................................................................... | 15 | 17 | 0.06 | 0.06 | 0.06 | - | - |
| CCGGCCCCTGCCTTCACCAGCCACCCCCCAGAGCCAAGAAGACGTCCAAGAGGTGGACCAGGGGAGGAACGTGGTCCCGGCGCTCCAGACCCAGGAGTCCCAGAGACTGGCAGAGCTCGAAGAGCCCGGAGTAGCCCAACCCGAAGACTGTCCGTTACCTCTCGTGGAATTACCTCCGAACTGGCGCACGCCACGTCCCTCCCTCTCCTTCCTAGCAGCGAGGGTGAACCGGGCCCCGAGGCCGCGGTTTCGCTCCCGGCCCCTCTGCCCCCATCGCCCCCTCCACCGCCCCCGCCGCCCCCTCCTCCACCCCCGCCTCTGCCCACGAAGGAGGACGGCGCGGAGCCGCT ...................................................................................................((((((((.(((((.((((...)))).(((.((......)))))....))))).)).....))).)))..........((..((((....))))..))......................................................................................................................................................... ...................................................................................................100.....................................................................................................203................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553592(SRX182798) source: Brain. (Brain) |
|---|