| (1) HEART | (1) KIDNEY | (1) OTHER |
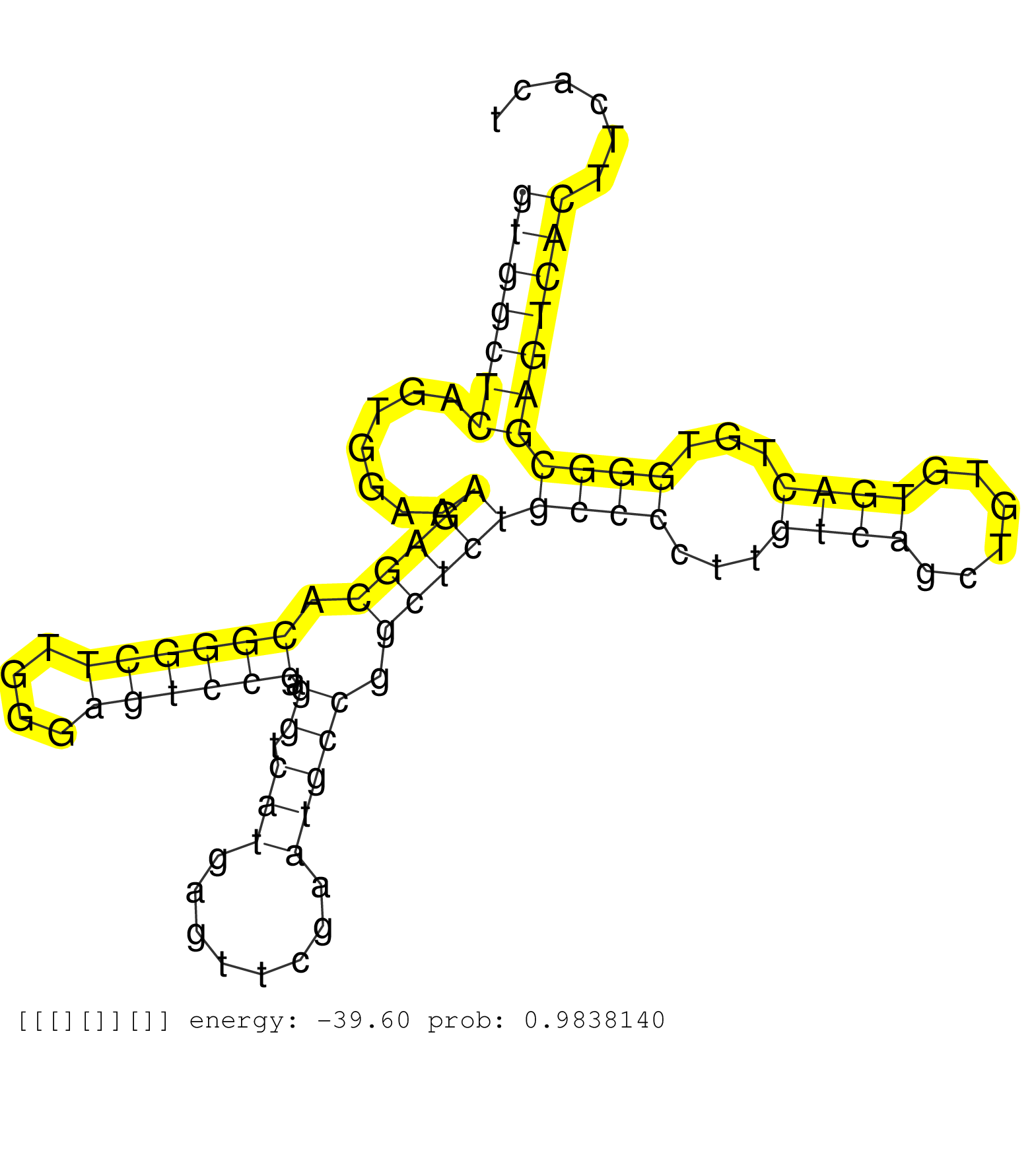
| GAAAGGACCCGTATCCTTTGAGAAATCACAACGATTTAGAAAGCAAAAAGGTAAGAAATCGCTTACAGTGAAAAAGATCGCATTTTGTGTGTTCTAATTTGATAAGACACTCCTGCGAATACCATTTCAGTGAAGACATGCTGCGGCAAACATGAGGTTTTGGCTGTCCTGAAAGCCTGAGAGTAGTGTTAATTGAGAAGCAGCGTGGCTCAGTGGAAAGAGCACGGGCTTGGGAGTCCGAGGTCATGAGTTCGAATGCCGGCTCTGCCCCTTGTCAGCTGTGTGACTGTGGGCGAGTCACTTCACTTCTCTGGGCCTCAGTGACCTCATCTGTAAAATGGGGATGAAGA ............................................................................................................................................................................................................(((((((.......(((((.((((((....)))))).((.(((........))))).)))))((((...((((......))))...)))))))))))................................................. ............................................................................................................................................................................................................205...................................................................................................307......................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................................................................................TCAGCTGTGTGACTGTGGGCGAGTAAT................................................. | 27 | 1.00 | 0.00 | 1.00 | - | - | |
| ..................................................................................................................................................................................................................................................................................TCAGCTGTGTGACTGTGGGCGAGTAATA................................................ | 28 | 1.00 | 0.00 | 1.00 | - | - | |
| .......................................................................................................................................................................................................................................................................................TGTGTGACTGTGGGCGAGTCAAAC............................................... | 24 | 1.00 | 0.00 | - | 1.00 | - | |
| ..................................................................................................................................................................................................................................................................................TCAGCTGTGTGACTGTGGGCGAGTTTAA................................................ | 28 | 1.00 | 0.00 | 1.00 | - | - | |
| ....................................................................................................................................................................................................................................................................................AGCTGTGTGACTGTGGGCGAGTATA................................................. | 25 | 1.00 | 0.00 | 1.00 | - | - | |
| ....................................................................................................................................................................................................................................................................................AGCTGTGTGACTGTGGGCGAGTAAAA................................................ | 26 | 1.00 | 0.00 | 1.00 | - | - | |
| .......................................................................................................................................................................................................................................................................................TGTGTGACTGTGGGCGAGTAAA................................................. | 22 | 1.00 | 0.00 | 1.00 | - | - | |
| .................................................................................................................................................................................................................TCAGTGGAAAGAGCACGGGCTTGTA.................................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | - | |
| .................................................................................................................................................................................................................TCAGTGGAAAGAGCACGGGCTTGTGTT.................................................................................................................. | 27 | 1.00 | 0.00 | 1.00 | - | - | |
| ..................................................................................................................................................................................................................................................................................TCAGCTGTGTGACTGTGGGCGAGTAAA................................................. | 27 | 1.00 | 0.00 | 1.00 | - | - | |
| .......................................................................................................................................................................CCTGAAAGCCTGAGA........................................................................................................................................................................ | 15 | 9 | 0.11 | 0.11 | - | - | 0.11 |
| GAAAGGACCCGTATCCTTTGAGAAATCACAACGATTTAGAAAGCAAAAAGGTAAGAAATCGCTTACAGTGAAAAAGATCGCATTTTGTGTGTTCTAATTTGATAAGACACTCCTGCGAATACCATTTCAGTGAAGACATGCTGCGGCAAACATGAGGTTTTGGCTGTCCTGAAAGCCTGAGAGTAGTGTTAATTGAGAAGCAGCGTGGCTCAGTGGAAAGAGCACGGGCTTGGGAGTCCGAGGTCATGAGTTCGAATGCCGGCTCTGCCCCTTGTCAGCTGTGTGACTGTGGGCGAGTCACTTCACTTCTCTGGGCCTCAGTGACCTCATCTGTAAAATGGGGATGAAGA ............................................................................................................................................................................................................(((((((.......(((((.((((((....)))))).((.(((........))))).)))))((((...((((......))))...)))))))))))................................................. ............................................................................................................................................................................................................205...................................................................................................307......................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................................................ccctGGCATTCGAACTTCCC...................................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | |
| ......................................................................................................................................................................................................................................ccgcTCGAACTCATGACCTCGGACTCGCC........................................................................................... | 29 | 1.00 | 0.00 | 1.00 | - | - |