| (1) HEART | (1) KIDNEY | (2) OTHER |
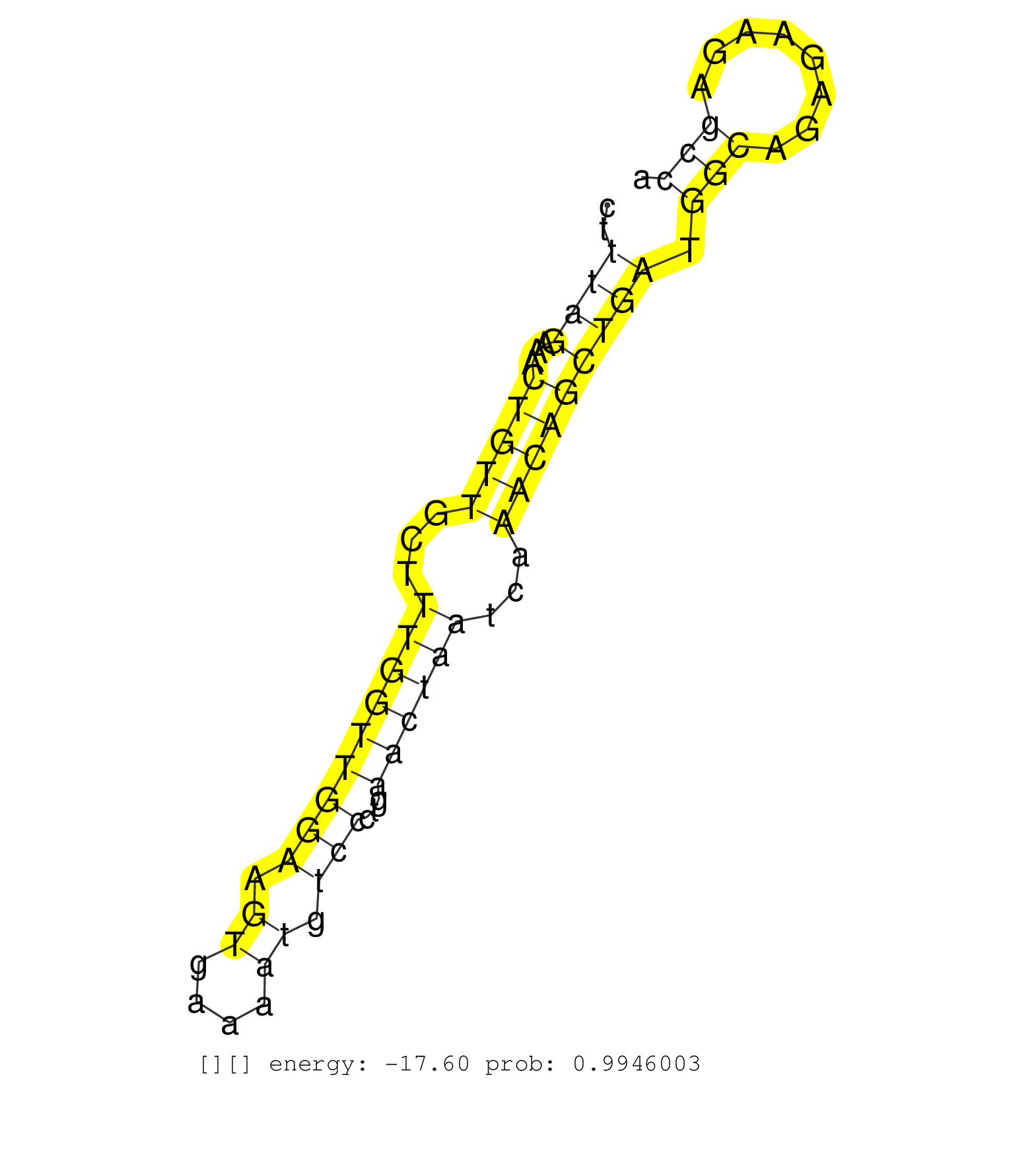
| CTTTAAGGATGGAGAAGCAAATAGAGCAGGAGTGGCTCATTTCCAATTAGATTATATTATTTGTCAAAGCTCTCTATTATGATACCCATAATTCTTGGCAGAAATAAAAGGAAAAAAGTTACAGGAGGTACTTTCTGAATAGAAGGAAATTGATATCTTAAGGGAAAAGTCAGAAAACATGTAAATGCTGGAGAAAAAGTTCATTCTGAATGATTTTTGCCATTTAATCATAGGCTTTAGAAACTGTTGCTTTGGTTGGAAGTGAAAATGTCCCTGAACTAATCAAACAGCTGATGGCAGAGAAGAGCCAGCTTCAAAATAATTTGCAGCACTGTCTTCATGAAATCCAA ............................................................................................................................................................................................................................................((((...(((((...(((((((((.((....)).)))...))))))...))))))))).(((........)))......................................... ..........................................................................................................................................................................................................................................235........................................................................310...................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................................GAAACTGTTGCTTTGGTTGGAAGT....................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..........................................................................................................................................................................................................................................................TTTGGTTGGAAGTGAAAATGTCC............................................................................. | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .......................................................................................................................................................................................................................................AGGCTTTAGAAACTGTTGCTTTGGTTGGAAGT....................................................................................... | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..................................................................................................................................................................................................................................................................................TGAACTAATCAAACAGCTGATGGCAG.................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...........................................................................................................................................................................................................................................................TTGGTTGGAAGTGAAAATGTCCC............................................................................ | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ........................................................................................................................................................................................................................................................................AAAATGTCCCTGAACTAATCAAACAGCTGATG...................................................... | 32 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .............................................................................................................................................................................................................................................................................................AACAGCTGATGGCAGAGATGC............................................ | 21 | 1.00 | 0.00 | - | 1.00 | - | - | |
| .................................................................................................................................................................................................................................................................................CTGAACTAATCAAACAGCTGATGGCAGAG................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| .............GAAGCAAATAGAGCA.................................................................................................................................................................................................................................................................................................................................. | 15 | 6 | 0.17 | 0.17 | 0.17 | - | - | - |
| CTTTAAGGATGGAGAAGCAAATAGAGCAGGAGTGGCTCATTTCCAATTAGATTATATTATTTGTCAAAGCTCTCTATTATGATACCCATAATTCTTGGCAGAAATAAAAGGAAAAAAGTTACAGGAGGTACTTTCTGAATAGAAGGAAATTGATATCTTAAGGGAAAAGTCAGAAAACATGTAAATGCTGGAGAAAAAGTTCATTCTGAATGATTTTTGCCATTTAATCATAGGCTTTAGAAACTGTTGCTTTGGTTGGAAGTGAAAATGTCCCTGAACTAATCAAACAGCTGATGGCAGAGAAGAGCCAGCTTCAAAATAATTTGCAGCACTGTCTTCATGAAATCCAA ............................................................................................................................................................................................................................................((((...(((((...(((((((((.((....)).)))...))))))...))))))))).(((........)))......................................... ..........................................................................................................................................................................................................................................235........................................................................310...................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................................................................................................TGAAGCTGGCTCTTCTCTGC.................................. | 20 | 2 | 1.00 | 1.00 | - | - | 1.00 | - |
| ........................................................................................................................................................................................................................................................................................................aggaTGAAGCTGGCTCTTCTAGGA.............................. | 24 | 0.50 | 0.00 | - | - | 0.50 | - | |
| ........................................................................................................................................................................................................................................................................................................agaTGAAGCTGGCTCTTCTCAGA............................... | 23 | 0.50 | 0.00 | - | - | 0.50 | - |