| (1) HEART | (1) KIDNEY | (1) OTHER |
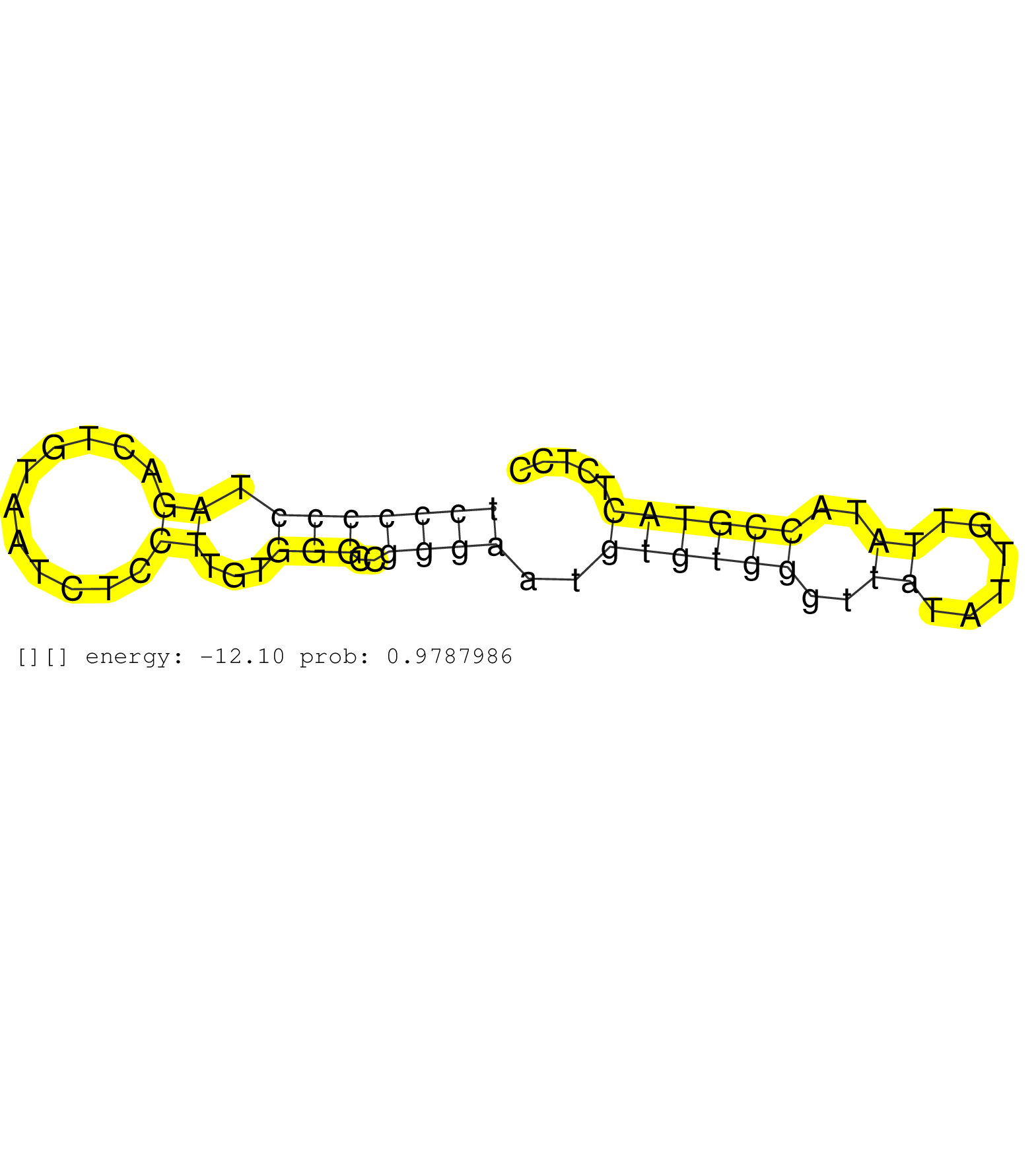
| CCCACAGCATTAACATACATATCCGCAGTTTATTGACGTTAATGTCTGTCTCCCCCCTAGACTGTAATCTCCTTGTGGGCCGGGAATGTGTGGGTTATATTGTTATACCGTACTCTCCCAAGCACTTAGTCCAGTGTTCTGCATACAGTAAGTGTTCAATAAATGTGATTGTTAGATTGATTGAAGTGGCAGGACAGGGATTAGAACCCAGACCTCTGACTCCCAGGCTCATGCACTTGCCATGCTGCTTCCCAAAATAACTAAACCTACCCTAACACTATTTTTGTGCTCTTCTTCTAGTTTGTGTTTCTGGCTTTTTGGGCCCTCTATCTGTACAACAGAGAGCTGAT ..................................................(((((((.((...........))...)))..))))..((((((..((......))..))))))............................................................................................................................................................................................................................................. ..................................................51.................................................................118...................................................................................................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| .........................................................TAGACTGTAATCTCCTTGTGGAAG............................................................................................................................................................................................................................................................................. | 24 | 3.00 | 0.00 | 3.00 | - | - | |
| .................................................................................................TATTGTTATACCGTACTCTCCCAAGCGA................................................................................................................................................................................................................................. | 28 | 2.00 | 0.00 | 2.00 | - | - | |
| .................................................................................................TATTGTTATACCGTACTCTCCCAT..................................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | |
| .................................................................................................TATTGTTATACCGTACTCTCCCAAAA................................................................................................................................................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | - | |
| .................................................................................................TATTGTTATACCGTACTCTCAAGC..................................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | |
| .........................................................TAGACTGTAATCTCCTTGTGGAAGG............................................................................................................................................................................................................................................................................ | 25 | 1.00 | 0.00 | 1.00 | - | - | |
| .........................................................TAGACTGTAATCTCCTTGTGCACA............................................................................................................................................................................................................................................................................. | 24 | 1.00 | 0.00 | 1.00 | - | - | |
| .................................................................................................TATTGTTATACCGTACTCTCCCAAGCAC................................................................................................................................................................................................................................. | 28 | 12 | 0.08 | 0.08 | 0.08 | - | - |
| ............................................................................................................................................................................................GCAGGACAGGGATTAGAACCC............................................................................................................................................. | 21 | 16 | 0.06 | 0.06 | - | - | 0.06 |
| CCCACAGCATTAACATACATATCCGCAGTTTATTGACGTTAATGTCTGTCTCCCCCCTAGACTGTAATCTCCTTGTGGGCCGGGAATGTGTGGGTTATATTGTTATACCGTACTCTCCCAAGCACTTAGTCCAGTGTTCTGCATACAGTAAGTGTTCAATAAATGTGATTGTTAGATTGATTGAAGTGGCAGGACAGGGATTAGAACCCAGACCTCTGACTCCCAGGCTCATGCACTTGCCATGCTGCTTCCCAAAATAACTAAACCTACCCTAACACTATTTTTGTGCTCTTCTTCTAGTTTGTGTTTCTGGCTTTTTGGGCCCTCTATCTGTACAACAGAGAGCTGAT ..................................................(((((((.((...........))...)))..))))..((((((..((......))..))))))............................................................................................................................................................................................................................................. ..................................................51.................................................................118...................................................................................................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................................................................................GAAACACAAACTAGAAGAAGAGC........................................ | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ..........................................................................................................................................................................................................agTGGGAGTCAGAGGTCTGGGTTGA........................................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | - |