| (1) HEART | (1) KIDNEY | (2) OTHER |
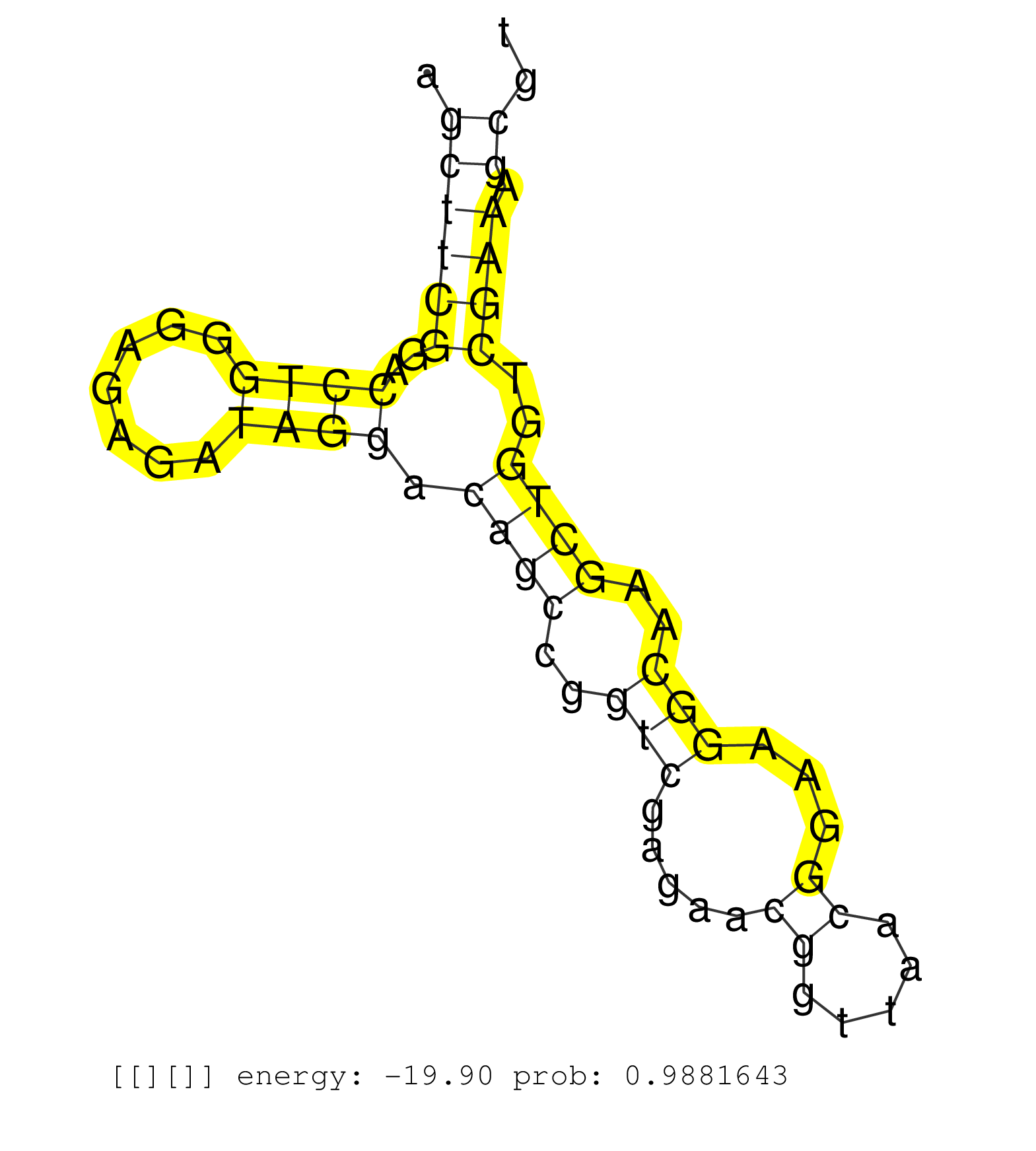
| GTACCTGGAAGAGCTCTGGGCGCAGGCCCGGTACATCTCCAACGCCGAAGGTCAGGGGCCGCTCCGCCCGACCGCCCCGTCGGCGGTCGGGCGCCCGCTTTGTGAGGAGCACTGTGCCGAGCGCGGGGGAGCGGGTAGACGCAACCCGTGCCCCGGGGGAGCTTCGGACCTGGGAGAGATAGGACAGCCGGTCGAGAACGGTTAACGGAAGGCAAGCTGGTCGAAAGCGTCGTAGGCGGCGGCGGCGGCAGCGATAGAGAAGCAGCGTGGCTCAGTGGAAAGAGCCCGGGCTTTGGAGTCAGGGCTCATGAGTTCGAATCCCAGCTCTGCCACCTGTCGGCTGTGTGACC ................................................................................................................................................................((((((..((((.......)))).((((..(((.....((.....))...)))..))))..)))).)).......................................................................................................................... ...............................................................................................................................................................160...................................................................230...................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................GGAAGGCAAGCTGGTCGAAA............................................................................................................................ | 20 | 1 | 2.00 | 2.00 | - | 2.00 | - | - |
| .......GAAGAGCTCTGGGCGC....................................................................................................................................................................................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| .........AGAGCTCTGGGCGCAGGCCCG................................................................................................................................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| .................................................................................................................................................................................................................................................................................................................TCATGAGTTCGAATCCCAGCTCTGAAA.................. | 27 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ...................................................................................................................................................................................................................................................................................................TTTGGAGTCAGGGCTCATGATTCC................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .....................................................................................................................................................................................GGACAGCCGGTCGAGAACGGTTAACGGAAGGC......................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| .............................................................................................................................................................................GAGAGATAGGACAGCCGGTCGAGAACGGTTAAC................................................................................................................................................ | 33 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ....................................................................................................................................................................CGGACCTGGGAGAGATAG........................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ...................................................................................................................................................................................................................................................................................................................ATGAGTTCGAATCCCAGCTAA...................... | 21 | 1.00 | 0.00 | 1.00 | - | - | - |
| GTACCTGGAAGAGCTCTGGGCGCAGGCCCGGTACATCTCCAACGCCGAAGGTCAGGGGCCGCTCCGCCCGACCGCCCCGTCGGCGGTCGGGCGCCCGCTTTGTGAGGAGCACTGTGCCGAGCGCGGGGGAGCGGGTAGACGCAACCCGTGCCCCGGGGGAGCTTCGGACCTGGGAGAGATAGGACAGCCGGTCGAGAACGGTTAACGGAAGGCAAGCTGGTCGAAAGCGTCGTAGGCGGCGGCGGCGGCAGCGATAGAGAAGCAGCGTGGCTCAGTGGAAAGAGCCCGGGCTTTGGAGTCAGGGCTCATGAGTTCGAATCCCAGCTCTGCCACCTGTCGGCTGTGTGACC ................................................................................................................................................................((((((..((((.......)))).((((..(((.....((.....))...)))..))))..)))).)).......................................................................................................................... ...............................................................................................................................................................160...................................................................230...................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................................................................................................................aaTTCGAACTCATGAGCCCTGAA.............................. | 23 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ........................................................................................................................................................................................................................................................................................................aaaaTTCGAACTCATGAGCCCTAAAA............................ | 26 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ................................................................................................................................................................................................................................................................................................................aaaaTGGGATTCGAACTCAAAAA....................... | 23 | 1.00 | 0.00 | 1.00 | - | - | - |