| (1) HEART | (1) KIDNEY | (2) OTHER |
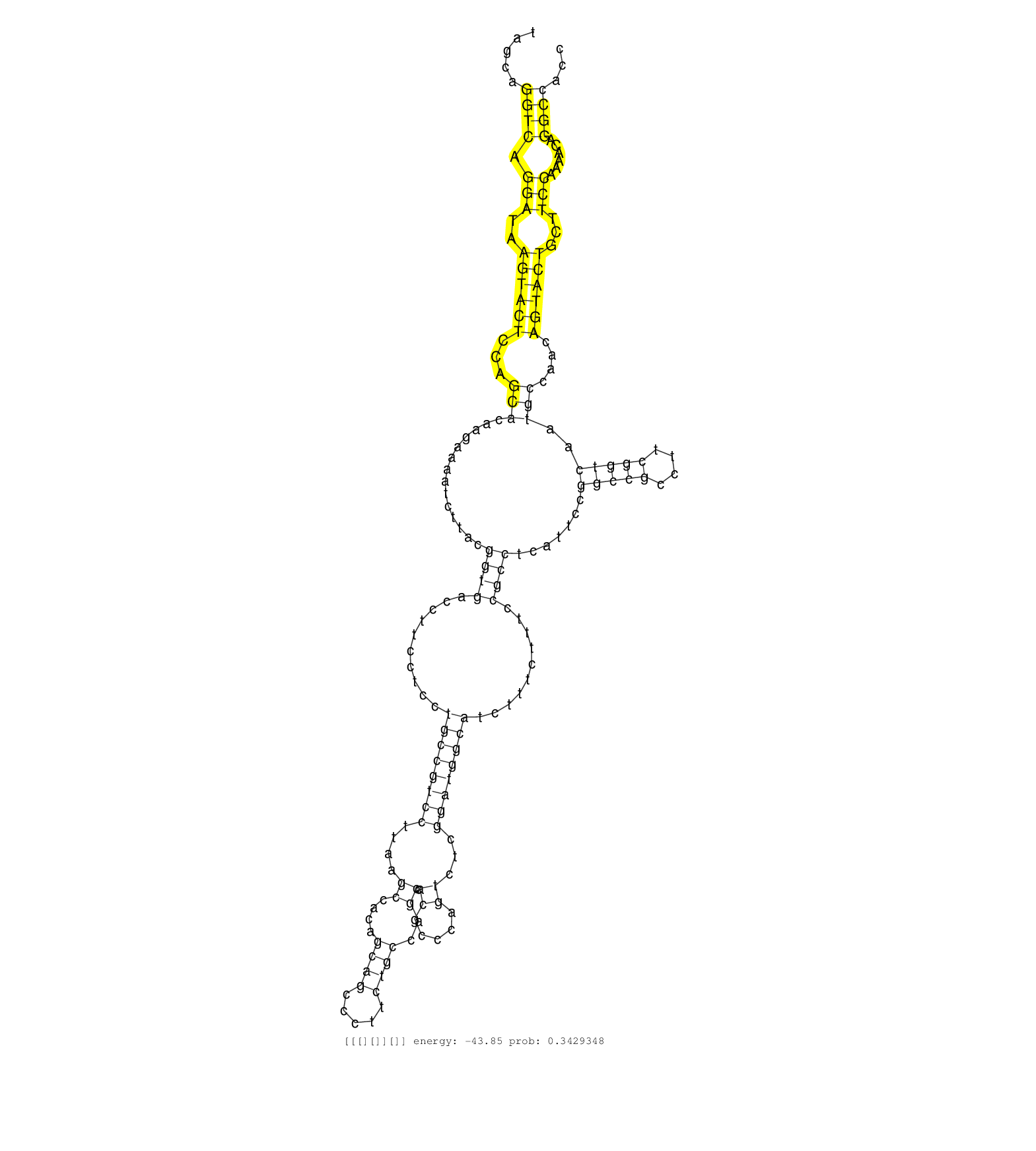
| CCTCTCCCCTGACACGATCTATACTGCCAAGTTTGTTGCGTACAATGATGATGAGGAGGAGGAGGAGGAGGAGGAGGAGGATTCCAGTCTAGCAGGTCAGGATAAGTACTCCAGCACAAGAAAATCTTACGGTGACCTTCCTCCTGCCGTCCTTAAGCCACAGCAGCCCTTCTGCCAGCCACGCCCAGTCTCGGATGGCATCTTTCTTTCCGCCTCATTCCGGCCGCCTTCGGTCAATGCCAACAGTACTGCTTCCAAAACAGGCCACCCCCCGCCCCCACCAAAAAAACCAAGTTCCTTCCATCCCAGCAACTGGAAAGGTAGAGCTCAAAACACGGCGAATGAAATTC ..............................................................................................((((.(((..((((((...(((..............((((..........((((((((....((....((((.....))))..)).((.....))...))))))))..........)))).......(((((....)))))..)))....))))))...)))......)))).................................................................................... .........................................................................................90.................................................................................................................................................................................269............................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|---|
| ......................................................................................GTCTAGCAGGTCAGGATAAGTACTCC.............................................................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - |
| ..............................................................................................GGTCAGGATAAGTACTCCAGC........................................................................................................................................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | 2.00 | - |
| .......................................................................................................................................................................................................................................................................................................................ACTGGAAAGGTAGAGCT...................... | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................CAGCAACTGGAAAGGTAGAGCTCAAAACACGG............ | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................AGCAACTGGAAAGGTAGAGCTCAAAACACGAC........... | 32 | 1.00 | 0.00 | - | - | - | 1.00 | |
| ....................................................................................................................................................................................................................................................................................................................GCAACTGGAAAGGTAGAGCTCAAAACACGGCG.......... | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .........TGACACGATCTATACTGCCAAGTTTGT.......................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..............................GTTTGTTGCGTACAATGATGATGAGG...................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ....................................................................................................................................................................................................................................................AGTACTGCTTCCAAAACAGGC..................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ..........................................................................................................................................................................................................................................................................................................................GGAAAGGTAGAGCTCAAAACACGGCGAATGA..... | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .....................................................................................AGTCTAGCAGGTCAGGATAAGTACTCC.............................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .....................................................................................................................................................................................................................................................................................................................CAACTGGAAAGGTAGAGCTCAAAACACGGC........... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................GTAGAGCTCAAAACACGGCG.......... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ....................................................................................................................................................................................ACGCCCAGTCTCGGA........................................................................................................................................................... | 15 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ..............................................................................................................................................................................................................................................................................................................................AGGTAGAGCTCAAAACACGGCGAATGAA.... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .......................................................................AGGAGGAGGATTCCAGTCTAGCAG............................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ........................................................................................................................................................................................................................................................................................................................CTGGAAAGGTAGAGC....................... | 15 | 8 | 0.25 | 0.25 | 0.25 | - | - | - |
| CCTCTCCCCTGACACGATCTATACTGCCAAGTTTGTTGCGTACAATGATGATGAGGAGGAGGAGGAGGAGGAGGAGGAGGATTCCAGTCTAGCAGGTCAGGATAAGTACTCCAGCACAAGAAAATCTTACGGTGACCTTCCTCCTGCCGTCCTTAAGCCACAGCAGCCCTTCTGCCAGCCACGCCCAGTCTCGGATGGCATCTTTCTTTCCGCCTCATTCCGGCCGCCTTCGGTCAATGCCAACAGTACTGCTTCCAAAACAGGCCACCCCCCGCCCCCACCAAAAAAACCAAGTTCCTTCCATCCCAGCAACTGGAAAGGTAGAGCTCAAAACACGGCGAATGAAATTC ..............................................................................................((((.(((..((((((...(((..............((((..........((((((((....((....((((.....))))..)).((.....))...))))))))..........)))).......(((((....)))))..)))....))))))...)))......)))).................................................................................... .........................................................................................90.................................................................................................................................................................................269............................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|