| (1) HEART | (1) KIDNEY | (1) OTHER |
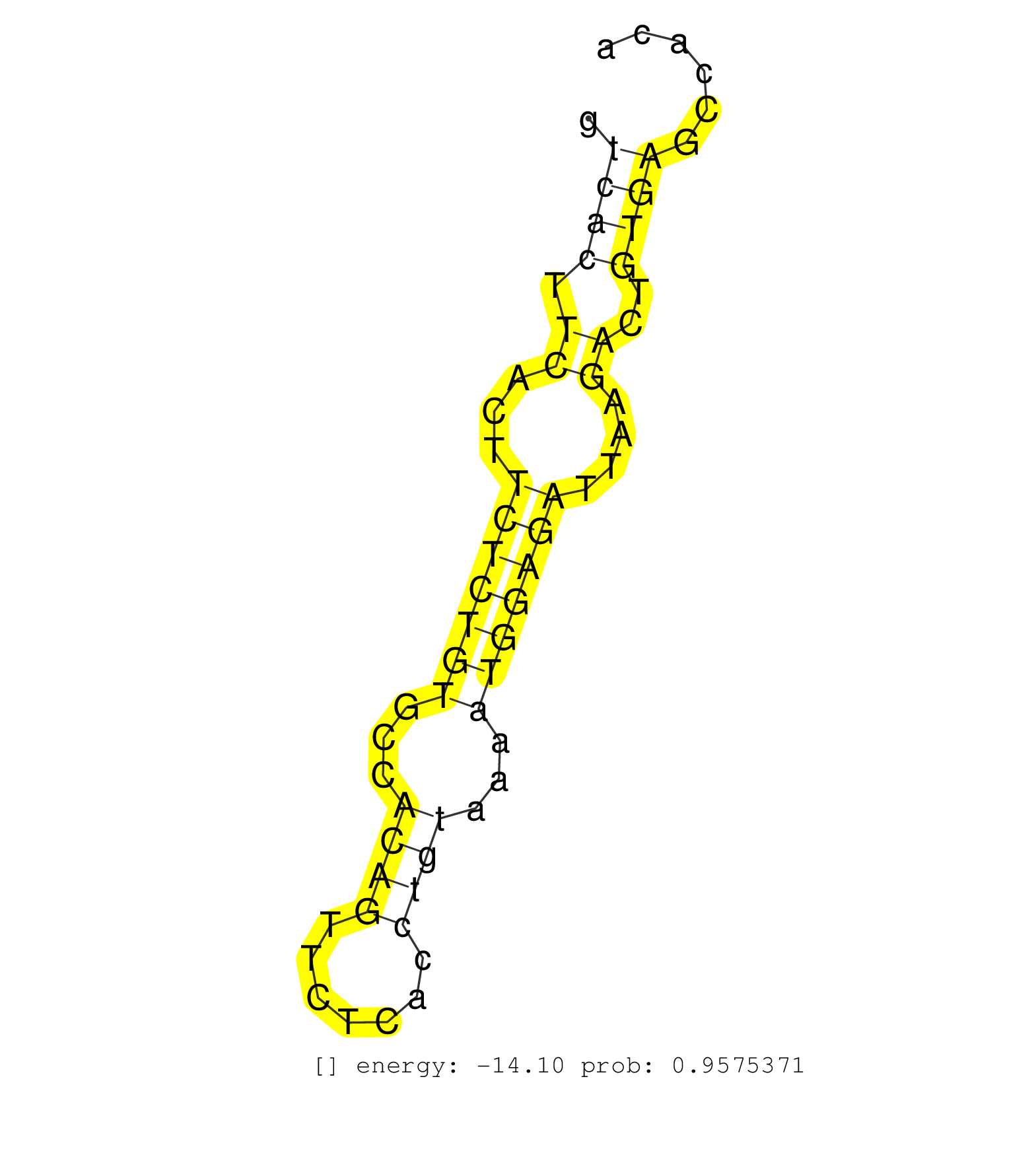
| GAAGAAGCTACCTCGAAGCCAATCGGTGTCTGCTGTAGAAGGCCTGAAAACTAAGCACCCTAGCTCCAAGGAAGCCAGAGGTACTGTGCGAGGCCGTCCAGCCAGAAGCAGCTGAAAGCAGTGTGGCCTAGTGGATAGAGCACAGGCATGGGAGTCAGAAGGACCTGGCTTCTCATCCCAATTCTGCAACATGCCTGCTGCATGACCTTGGGCGAGTCACTTCACTTCTCTGTGCCACAGTTCTCACCTGTAAAATGGAGATTAAGACTGTGAGCCACATGTGGGCATGGATAATGTCCATCCTGATTAGCTTATATCTACCCCAGCGCTTTAGAACAGTGTCTGGCACA ........................................................................................................................................................................................................................((((.((...(((((((...((((.......))))...)))))))....))..))))............................................................................. .......................................................................................................................................................................................................................216............................................................279..................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................................................TGGAGATTAAGACTGTTAAA........................................................................... | 20 | 37.00 | 0.00 | 37.00 | - | - | |
| ..................................GTAGAAGGCCTGAAAACTAAGCA..................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ...............................................................................................................................................................................................................................................................TGGAGATTAAGACTGTGCGAA.......................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | |
| ............................TCTGCTGTAGAAGGCCTGA............................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...............................................................................................................................................................................................................................................................TGGAGATTAAGACTGTGCAAT.......................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | |
| ...............................................................................................................................................................................................................................................................TGGAGATTAAGACTGTTATT........................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | |
| ........................................................................................................................................................................................................................................................TGTAAAATGGAGATTAAGACTGTTAAA........................................................................... | 27 | 1.00 | 0.00 | 1.00 | - | - | |
| ............................TCTGCTGTAGAAGGCCTGAAAACTAAGC...................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...............................................................................................................................................................................................................................................................TGGAGATTAAGACTGTGCAA........................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | |
| ............................................................................................................................................................................................................................TTCACTTCTCTGTGCCACAGTCAGC......................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | - | |
| .....................................................................................................................................................................................................................................................................TTAAGACTGTGAGCCACGTA..................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | |
| ...............................................................................................................................................................................................................................................................TGGAGATTAAGACTGTGAGCCAGAAA..................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | - | |
| .......................................AGGCCTGAAAACTAAAC...................................................................................................................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | 1.00 | |
| .................................TGTAGAAGGCCTGAAAACTAAGCACC................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| GAAGAAGCTACCTCGAAGCCAATCGGTGTCTGCTGTAGAAGGCCTGAAAACTAAGCACCCTAGCTCCAAGGAAGCCAGAGGTACTGTGCGAGGCCGTCCAGCCAGAAGCAGCTGAAAGCAGTGTGGCCTAGTGGATAGAGCACAGGCATGGGAGTCAGAAGGACCTGGCTTCTCATCCCAATTCTGCAACATGCCTGCTGCATGACCTTGGGCGAGTCACTTCACTTCTCTGTGCCACAGTTCTCACCTGTAAAATGGAGATTAAGACTGTGAGCCACATGTGGGCATGGATAATGTCCATCCTGATTAGCTTATATCTACCCCAGCGCTTTAGAACAGTGTCTGGCACA ........................................................................................................................................................................................................................((((.((...(((((((...((((.......))))...)))))))....))..))))............................................................................. .......................................................................................................................................................................................................................216............................................................279..................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................aaaAACTGTGGCACAGAGAAGTGAAAA......................................................................................................... | 27 | 1.00 | 0.00 | 1.00 | - | - |